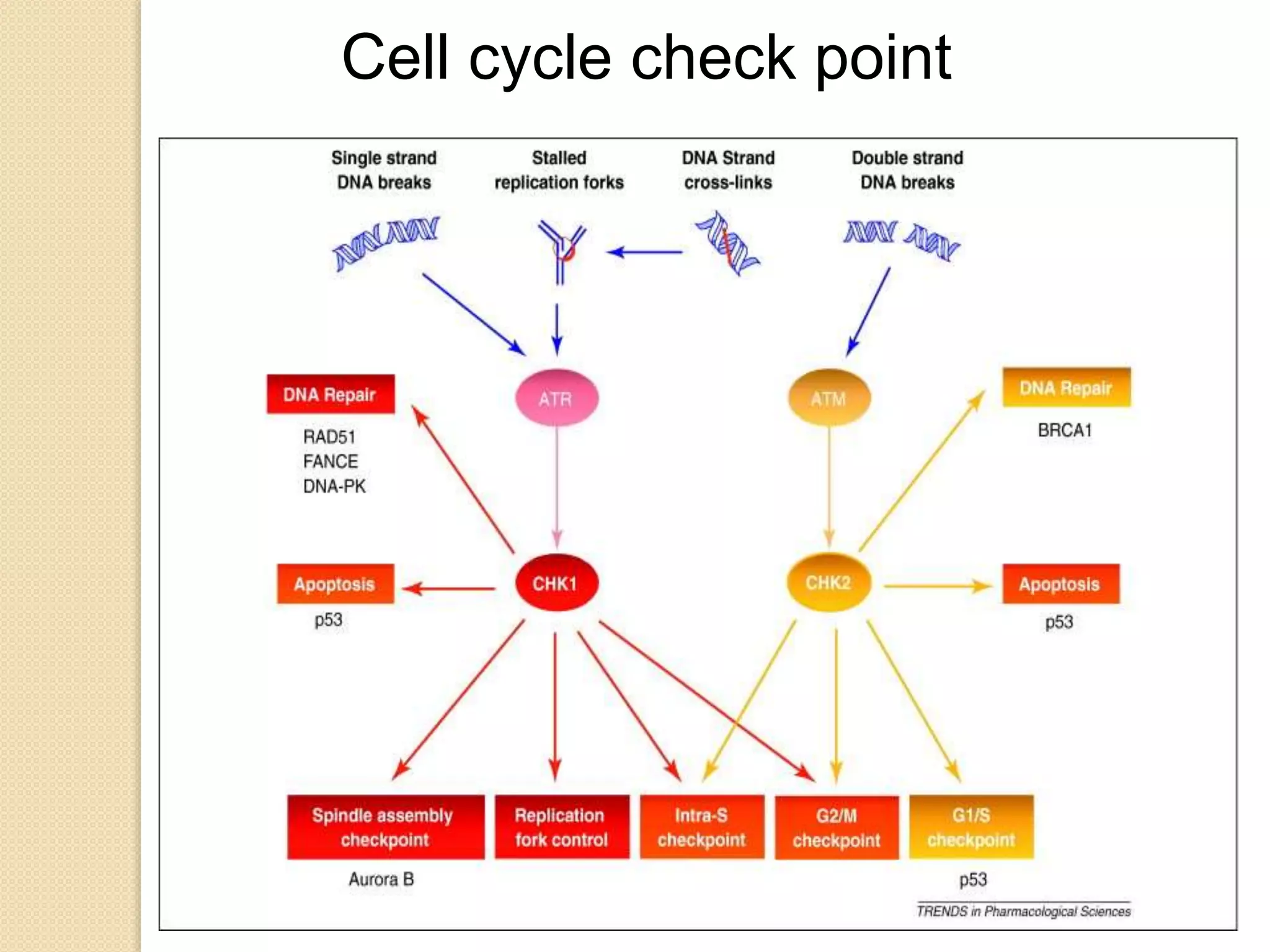
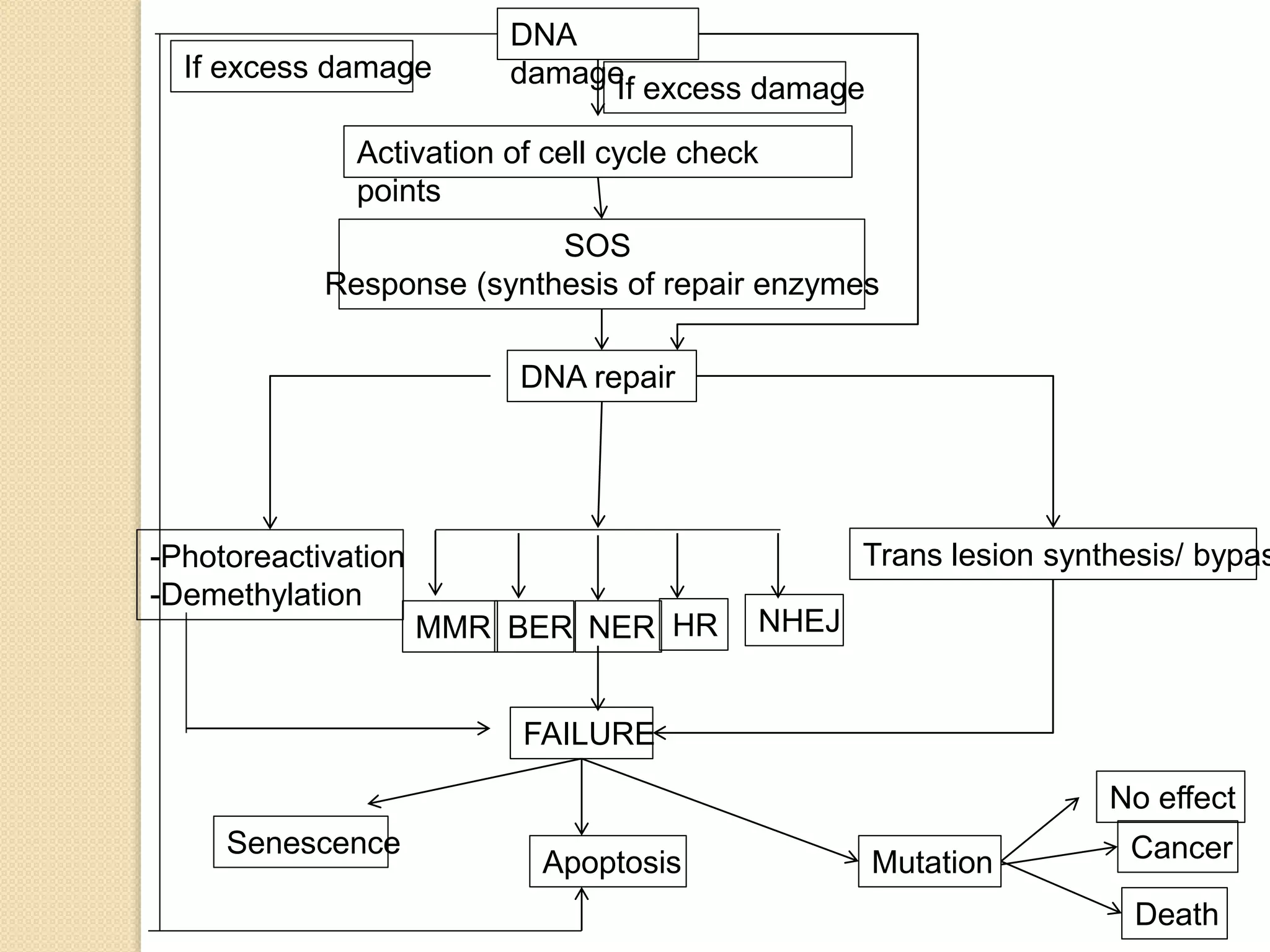
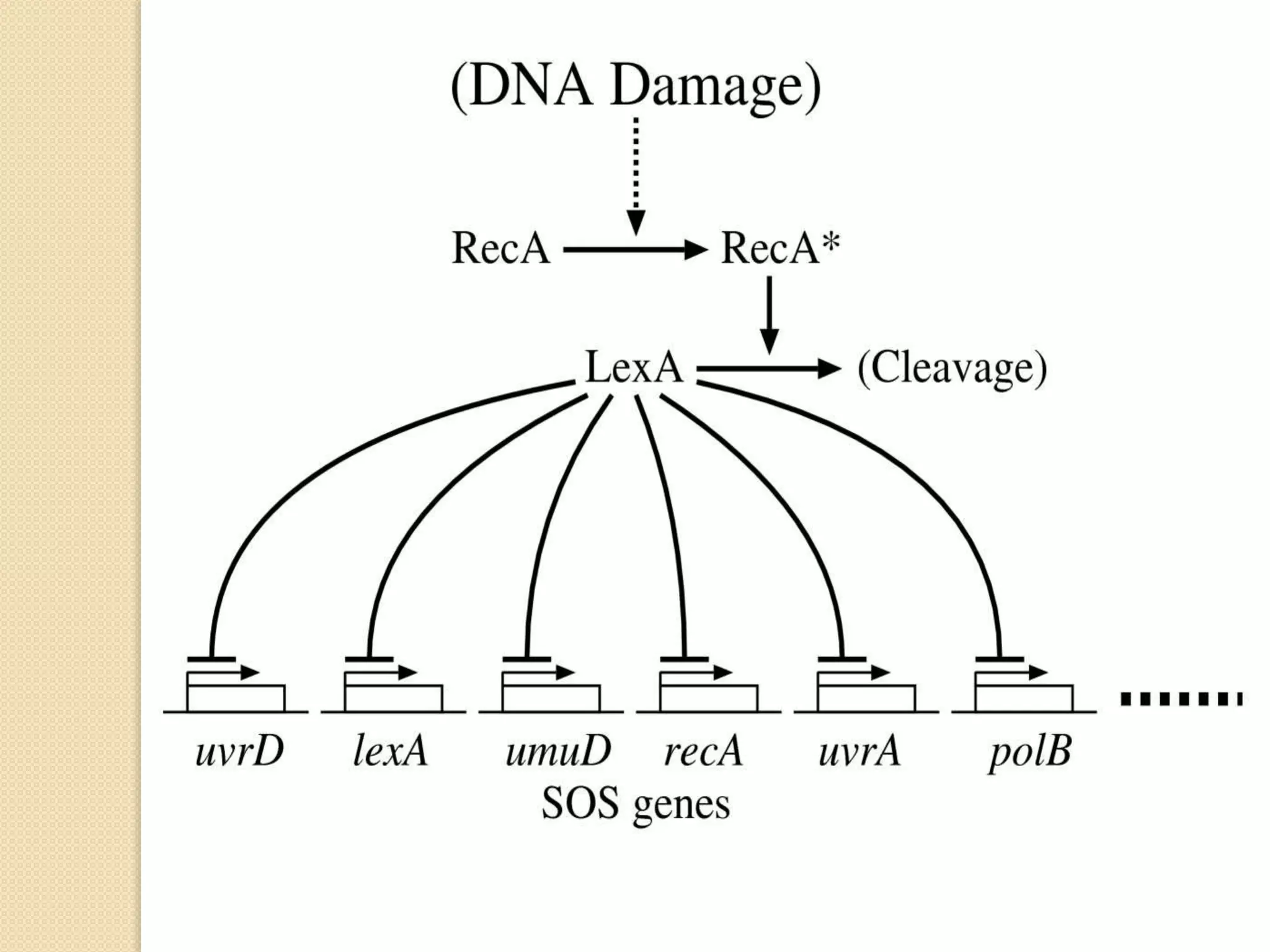
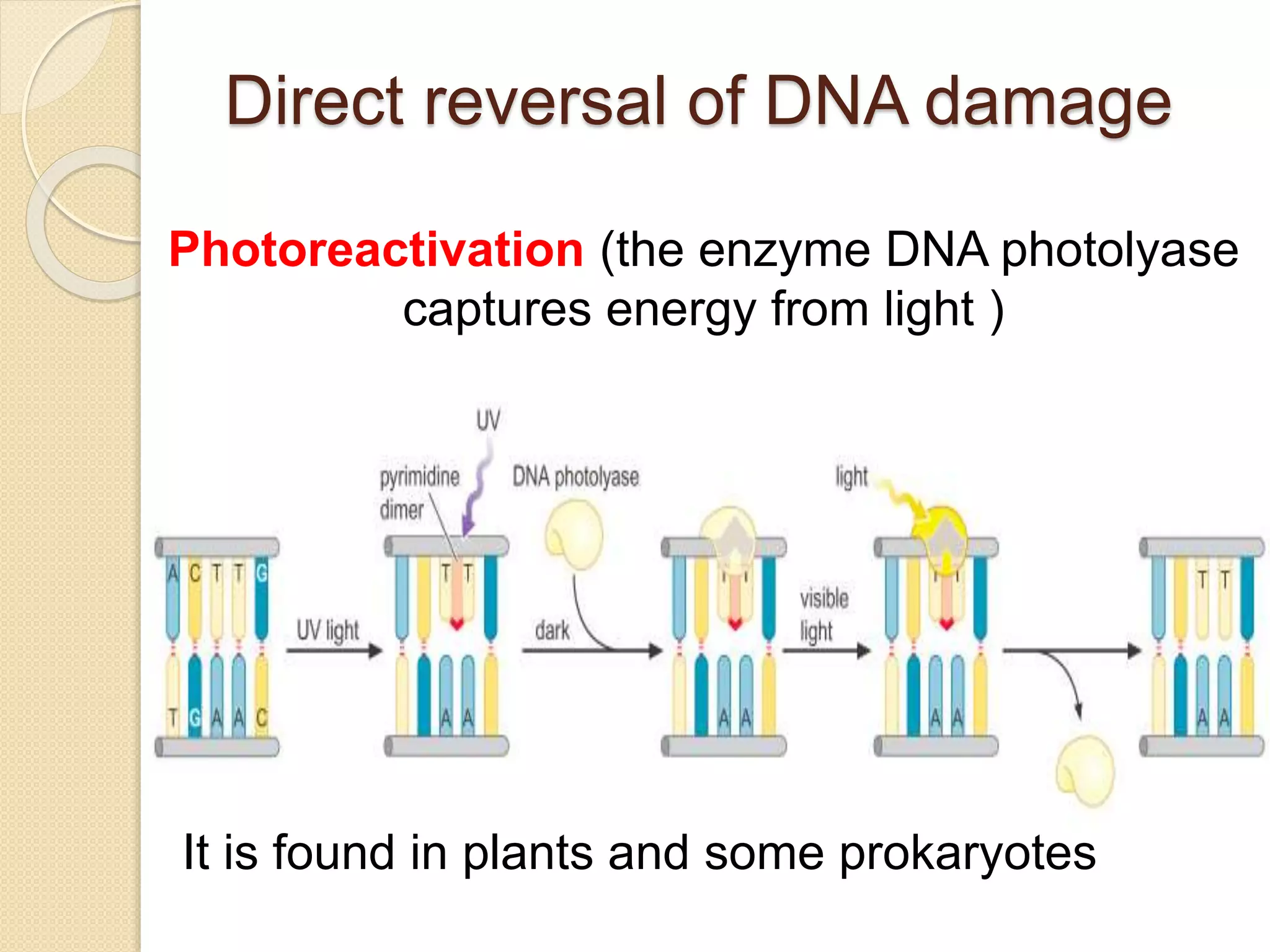
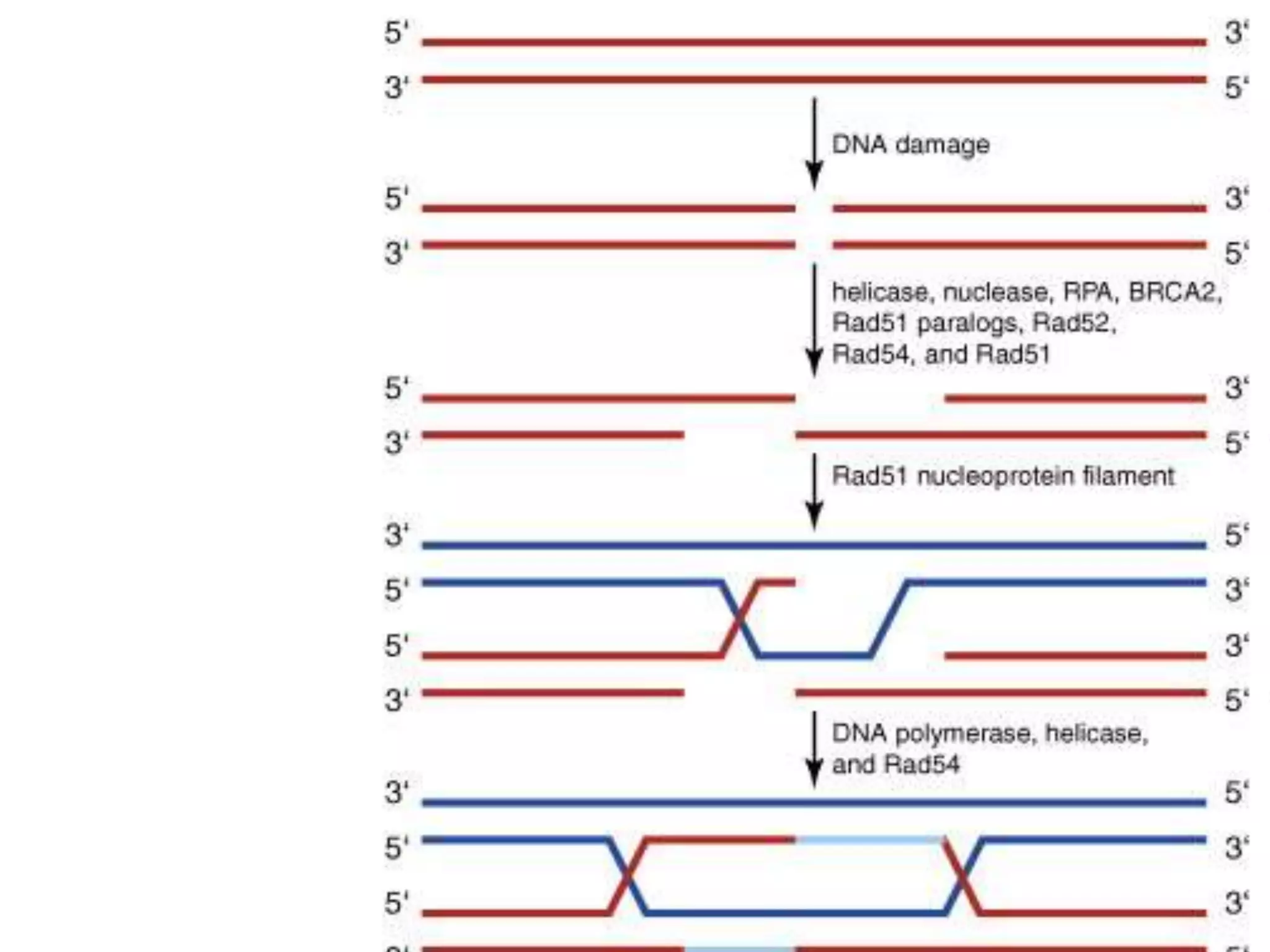
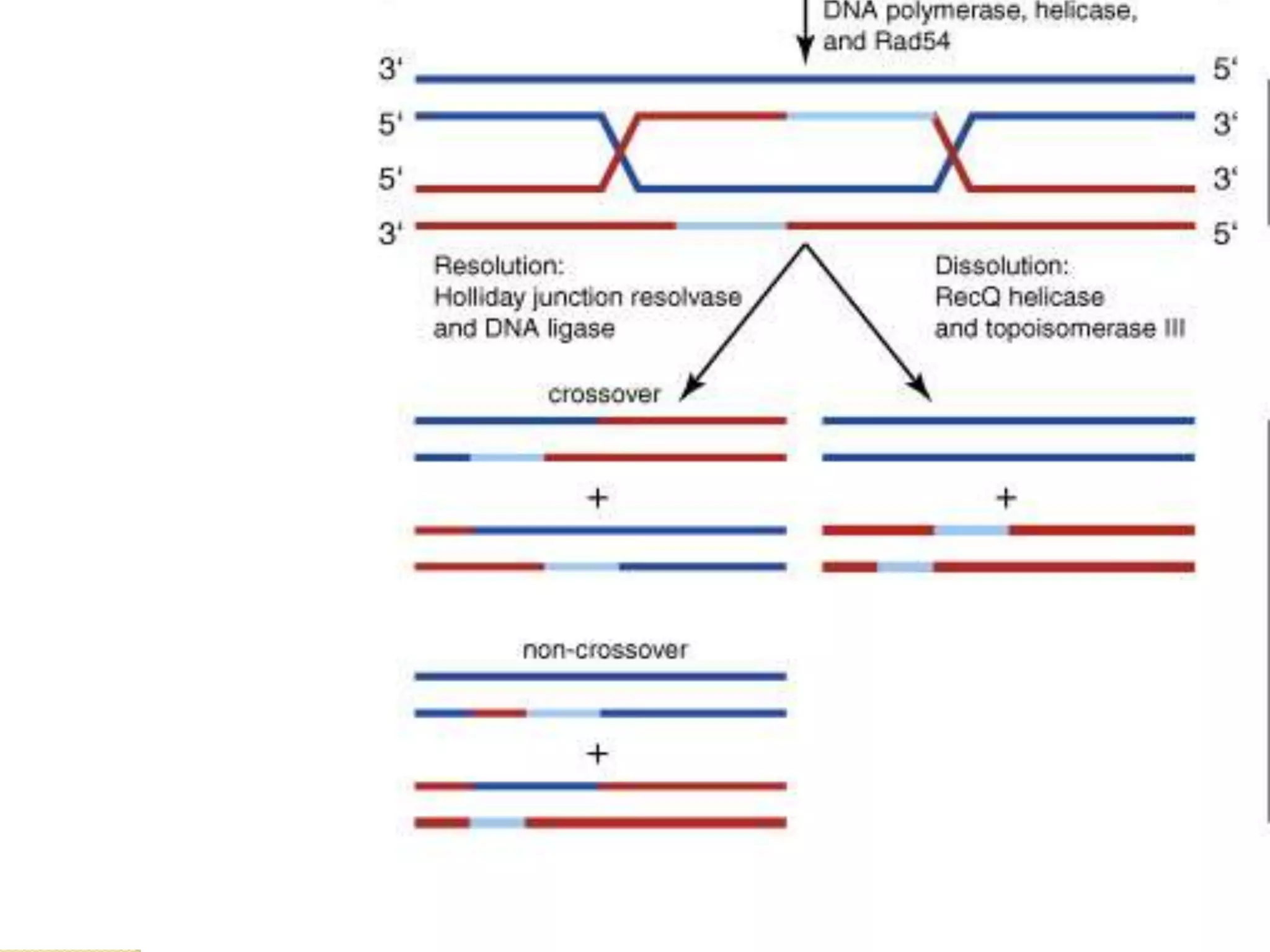
1. DNA repair is a collection of processes by which cells identify and correct damage to DNA molecules to maintain the integrity of the genome.
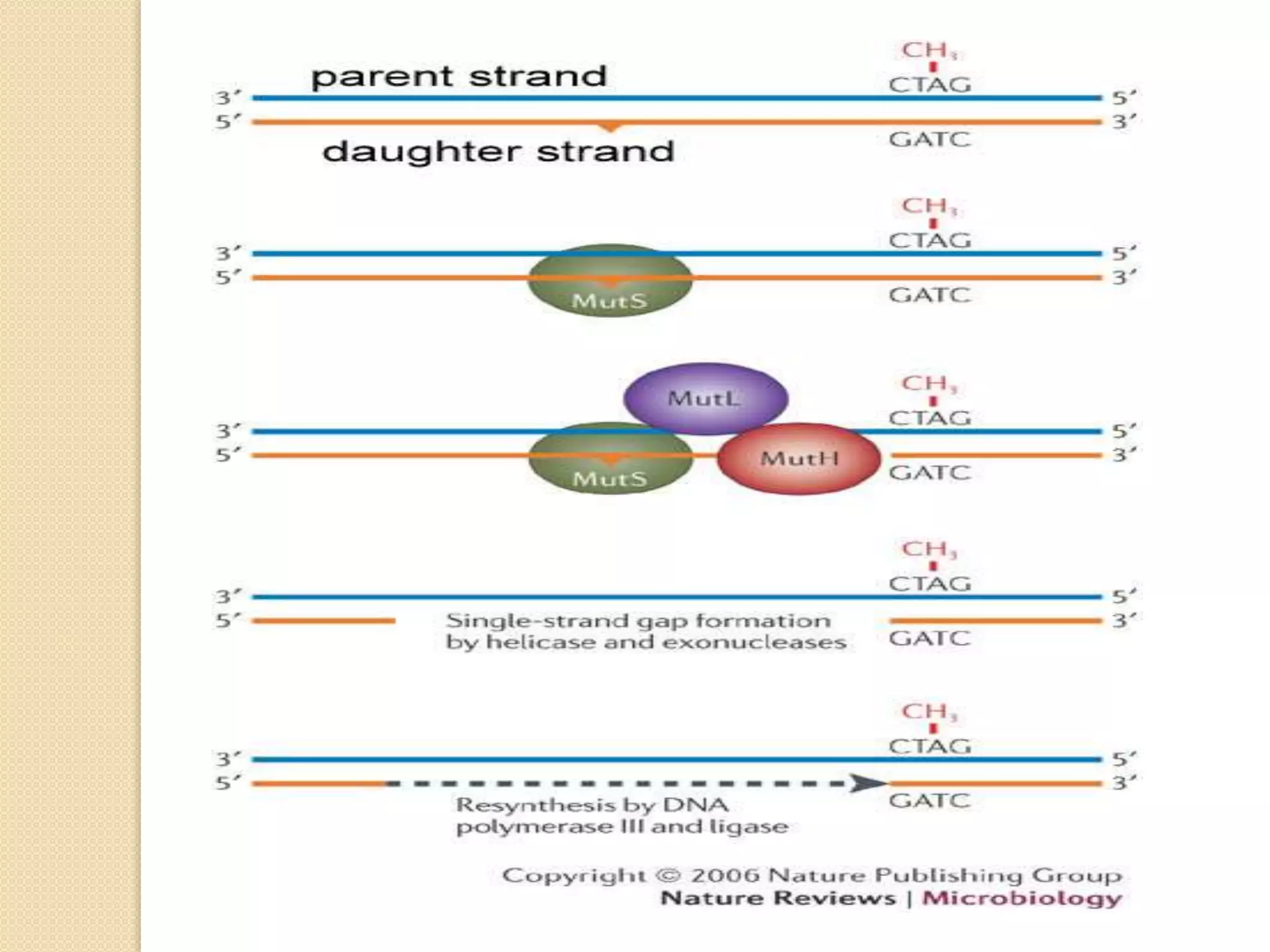
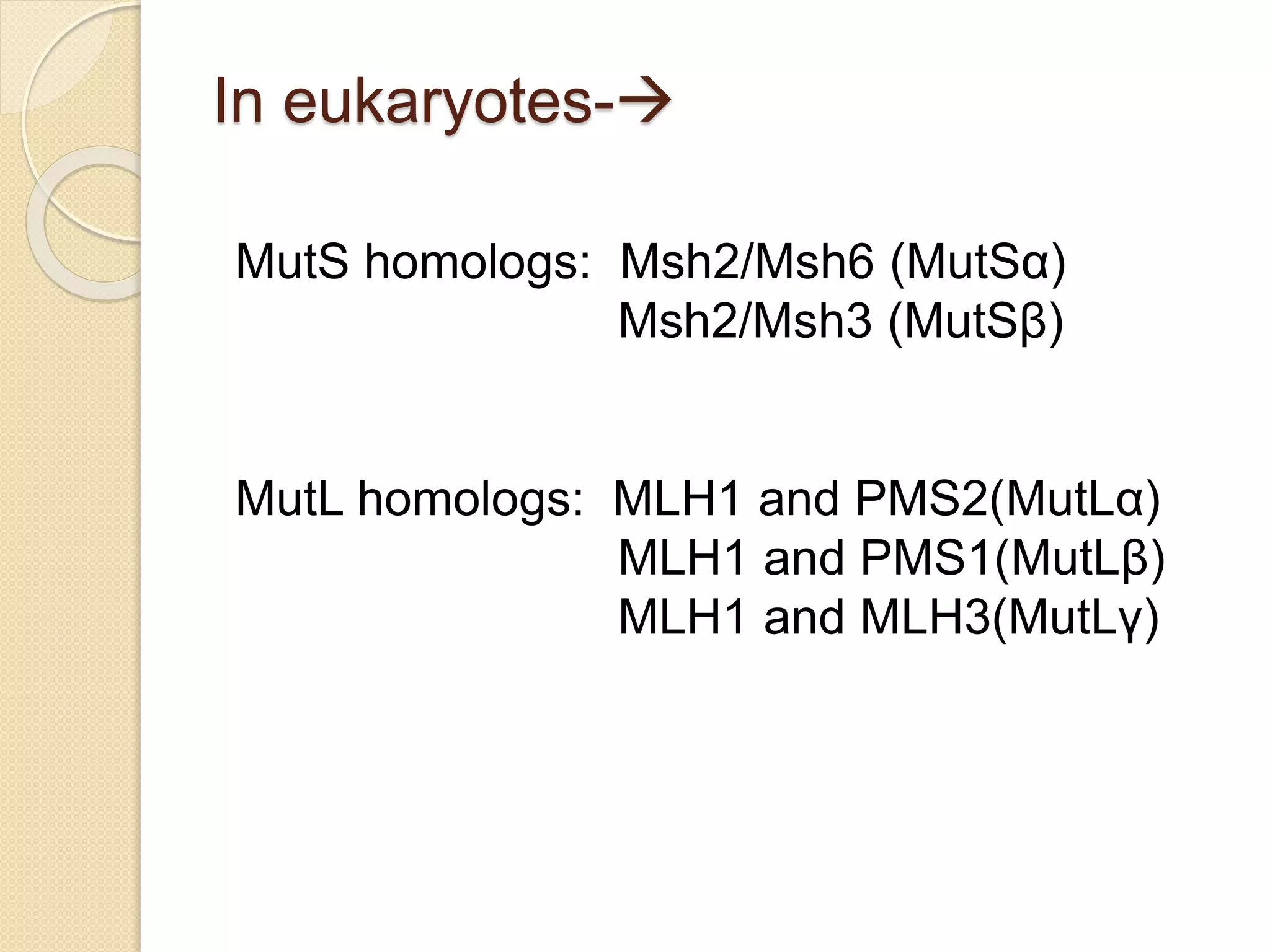
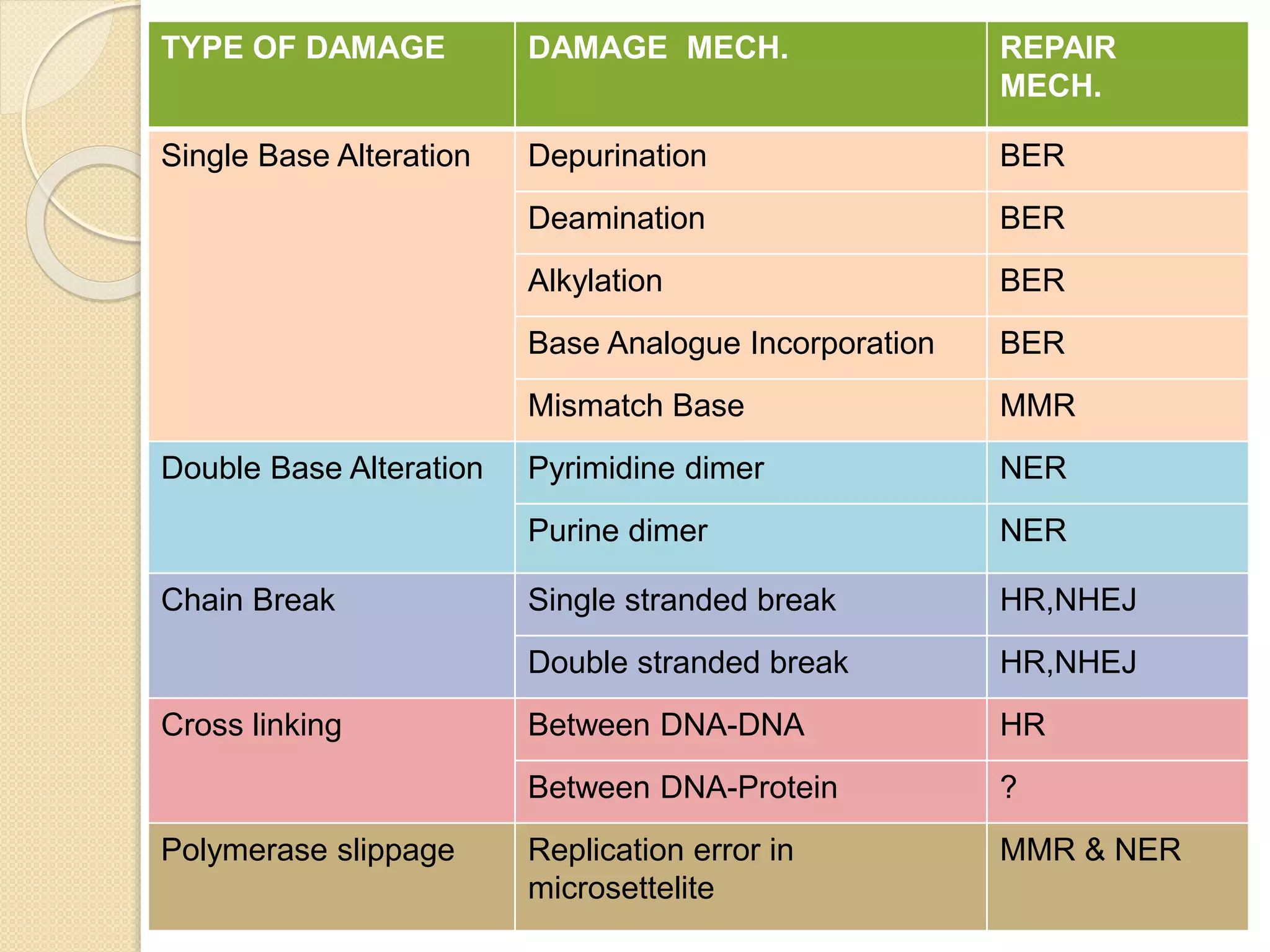
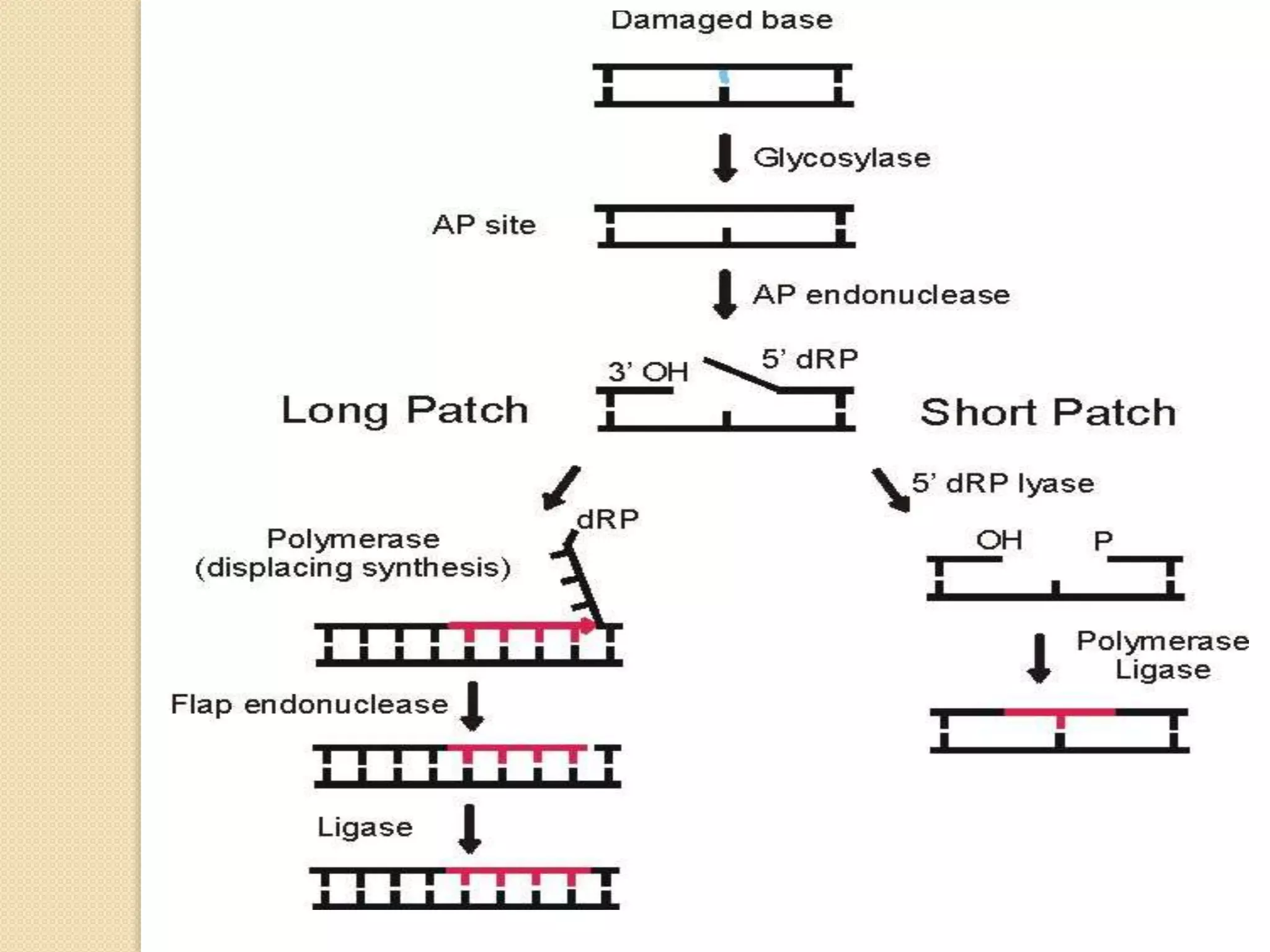
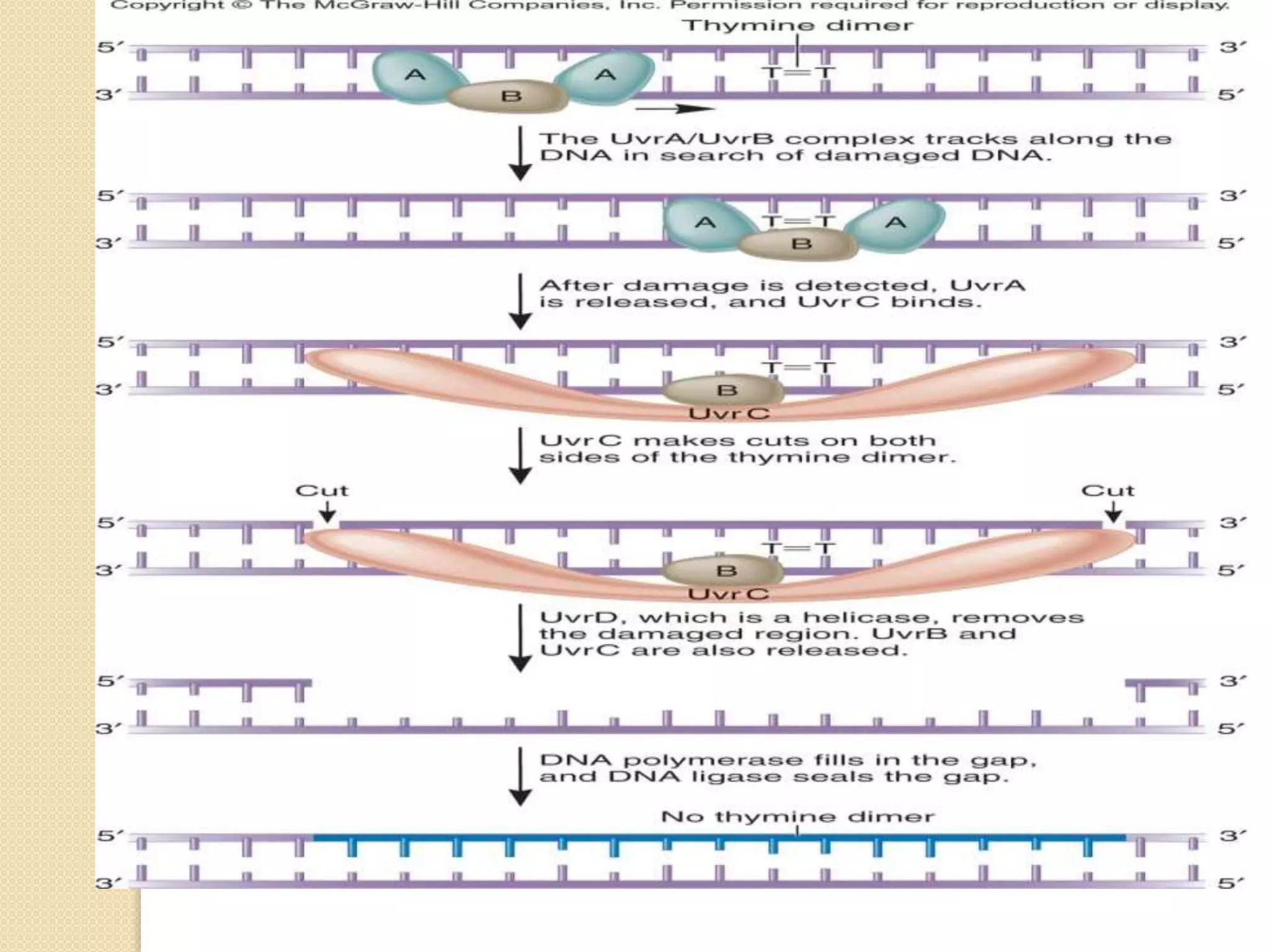
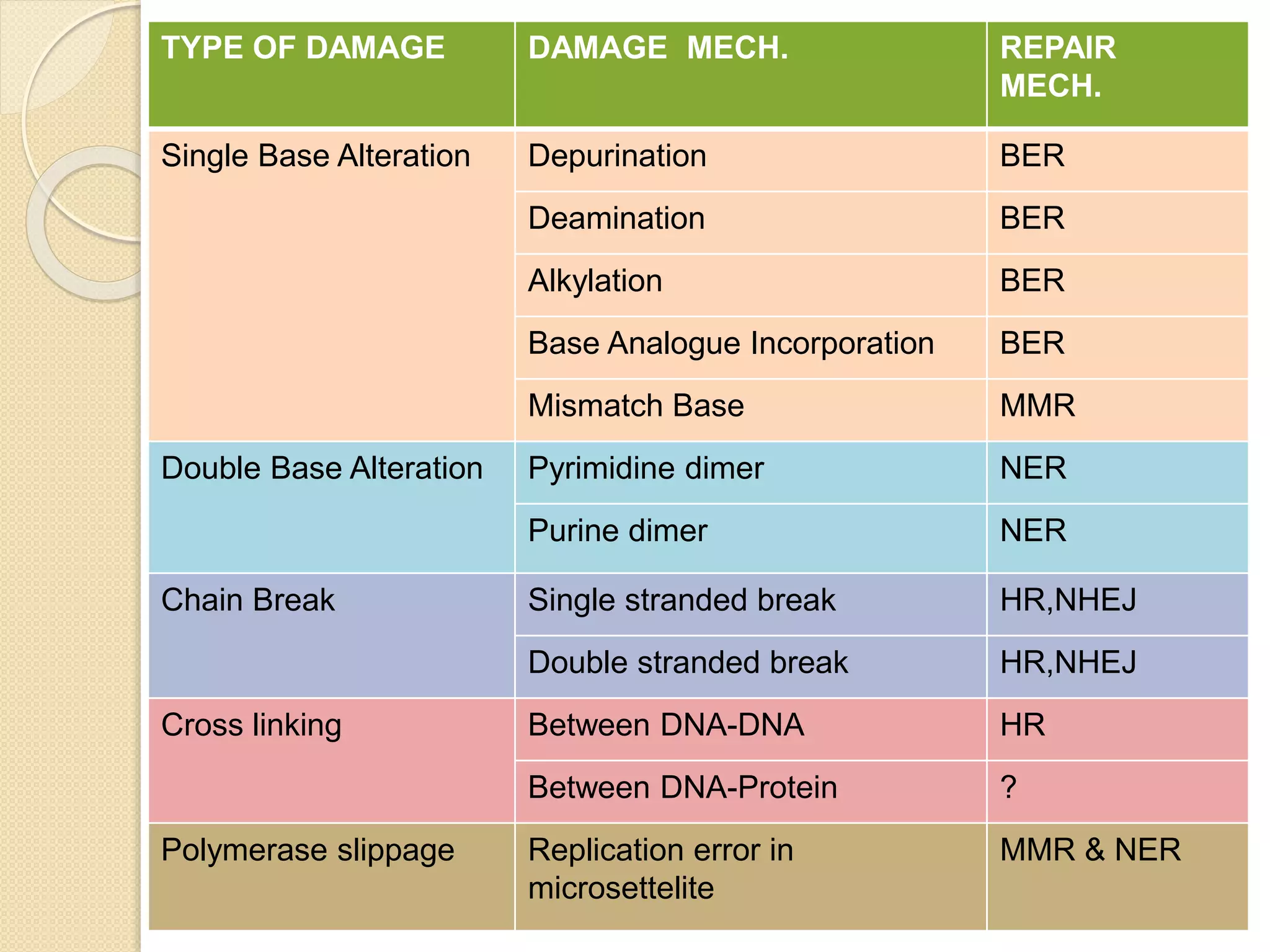
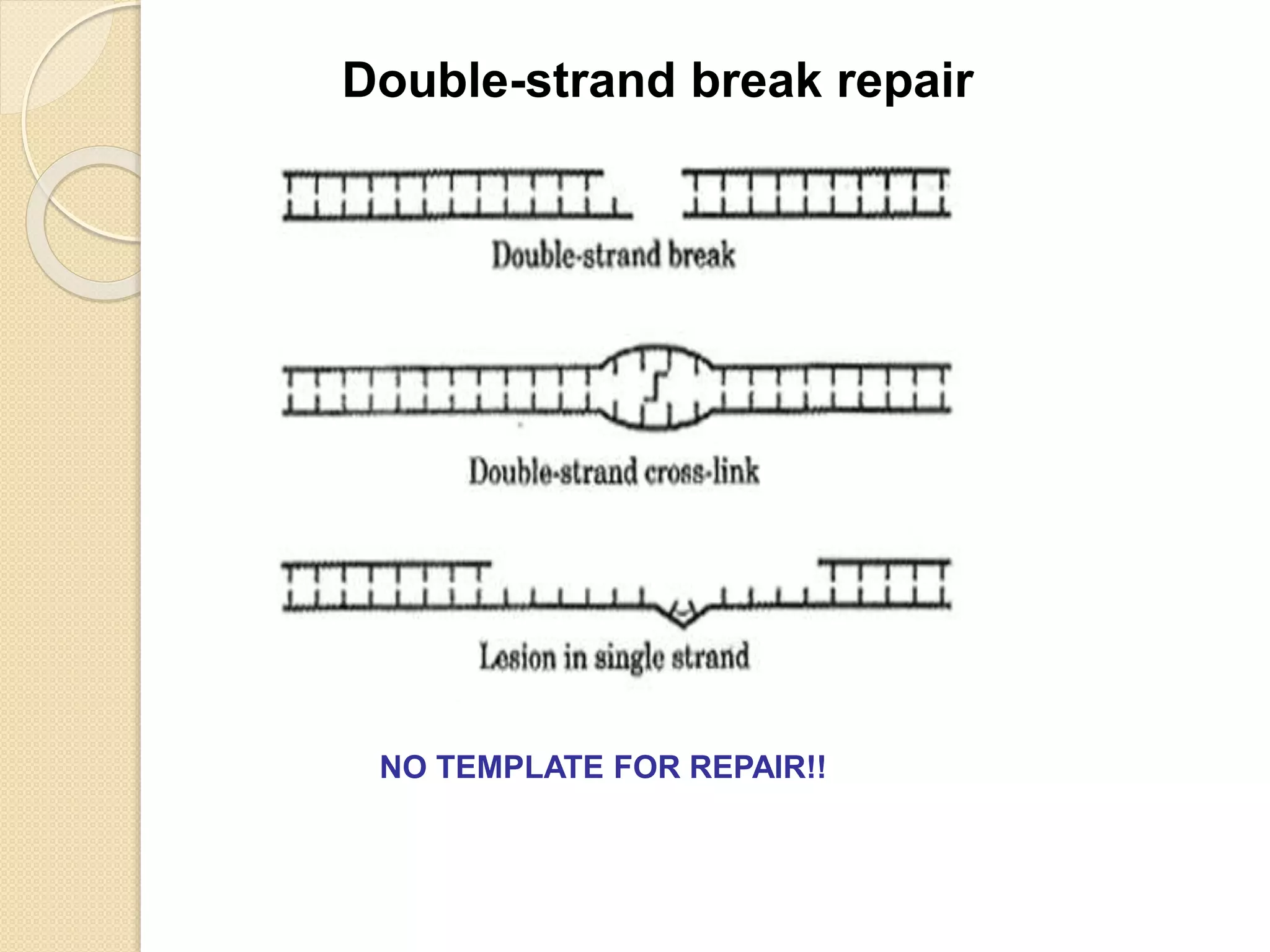
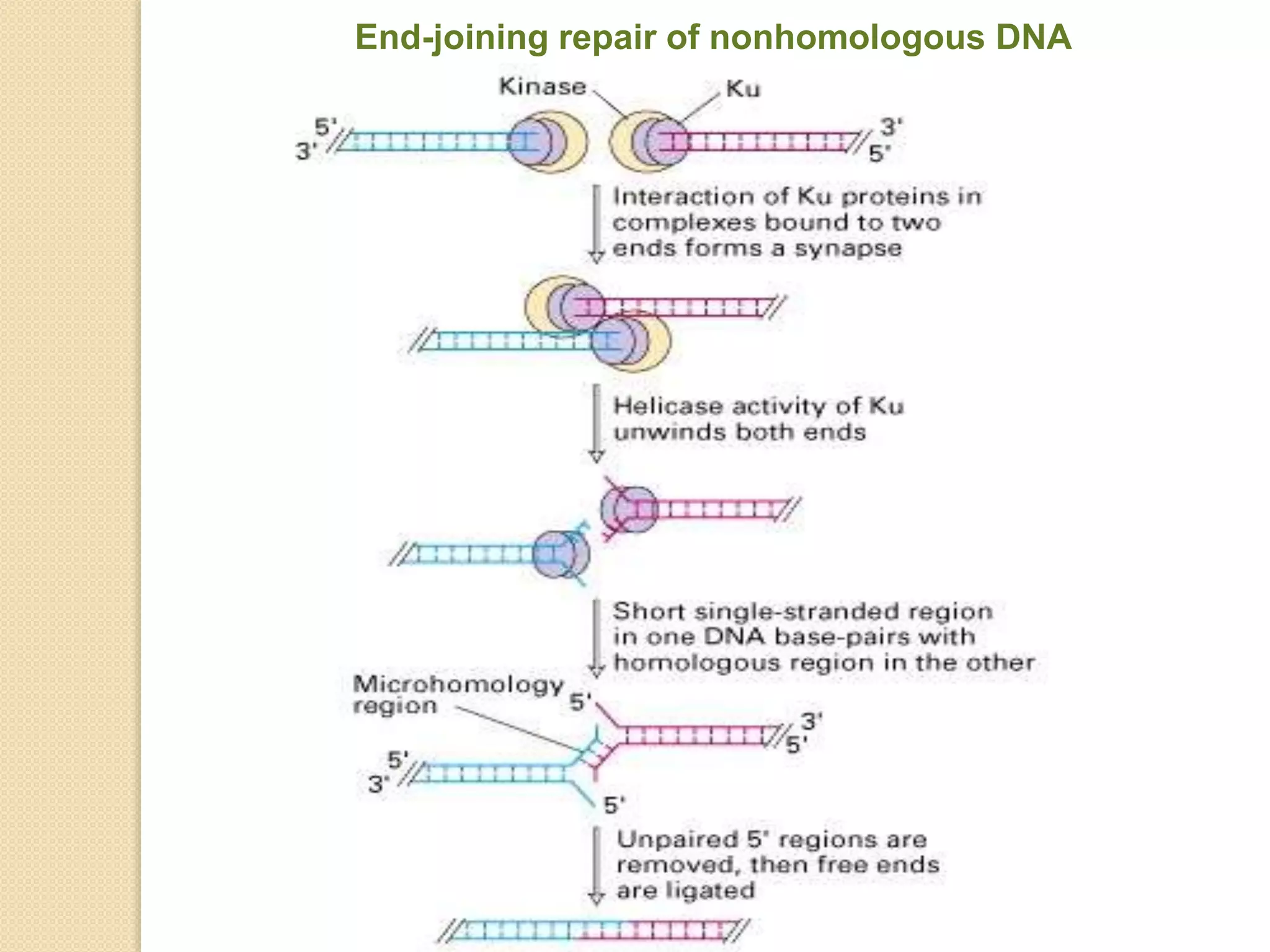
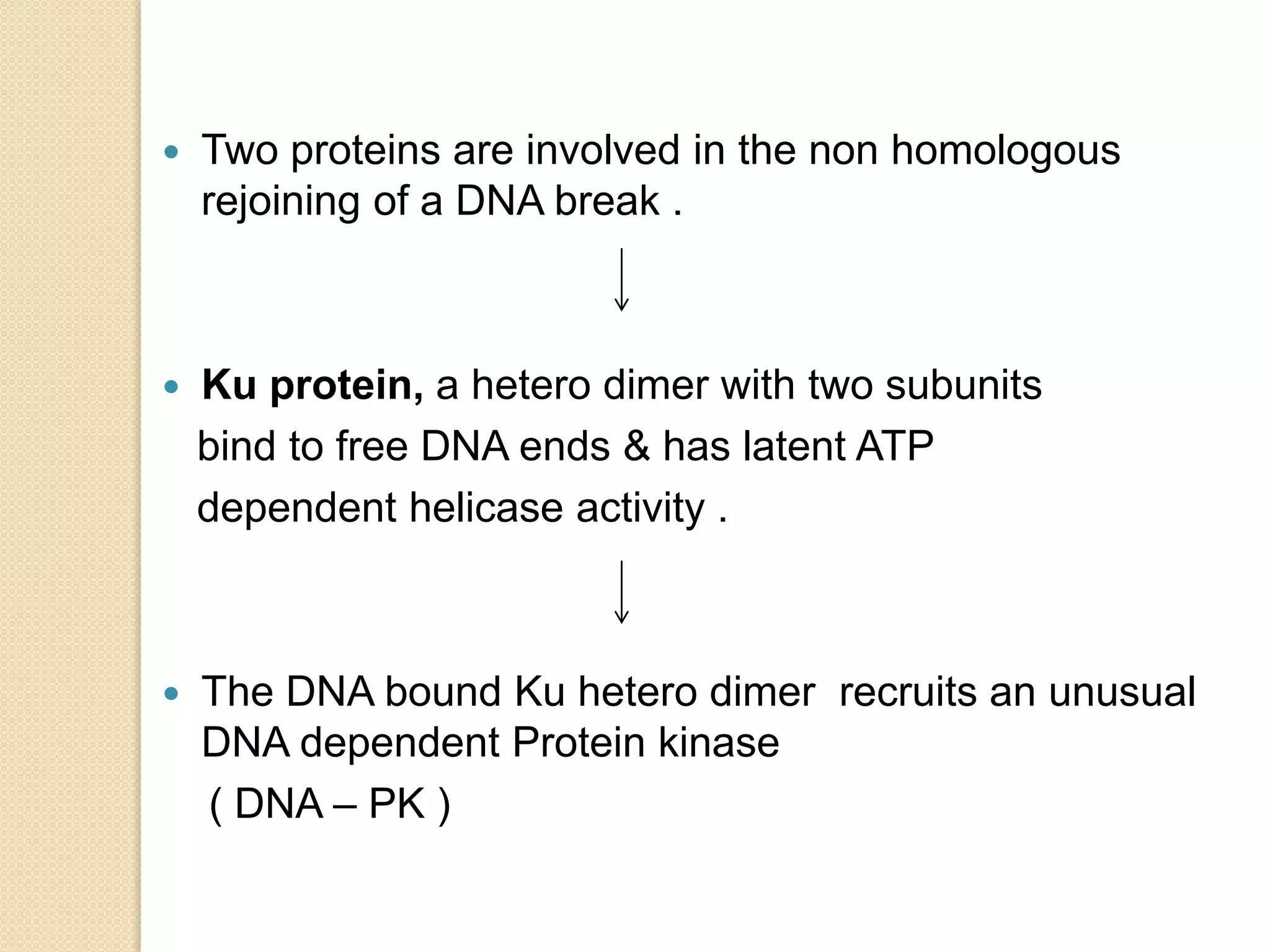
2. There are several pathways of DNA repair including base excision repair, nucleotide excision repair, mismatch repair, non-homologous end joining, and homologous recombination.
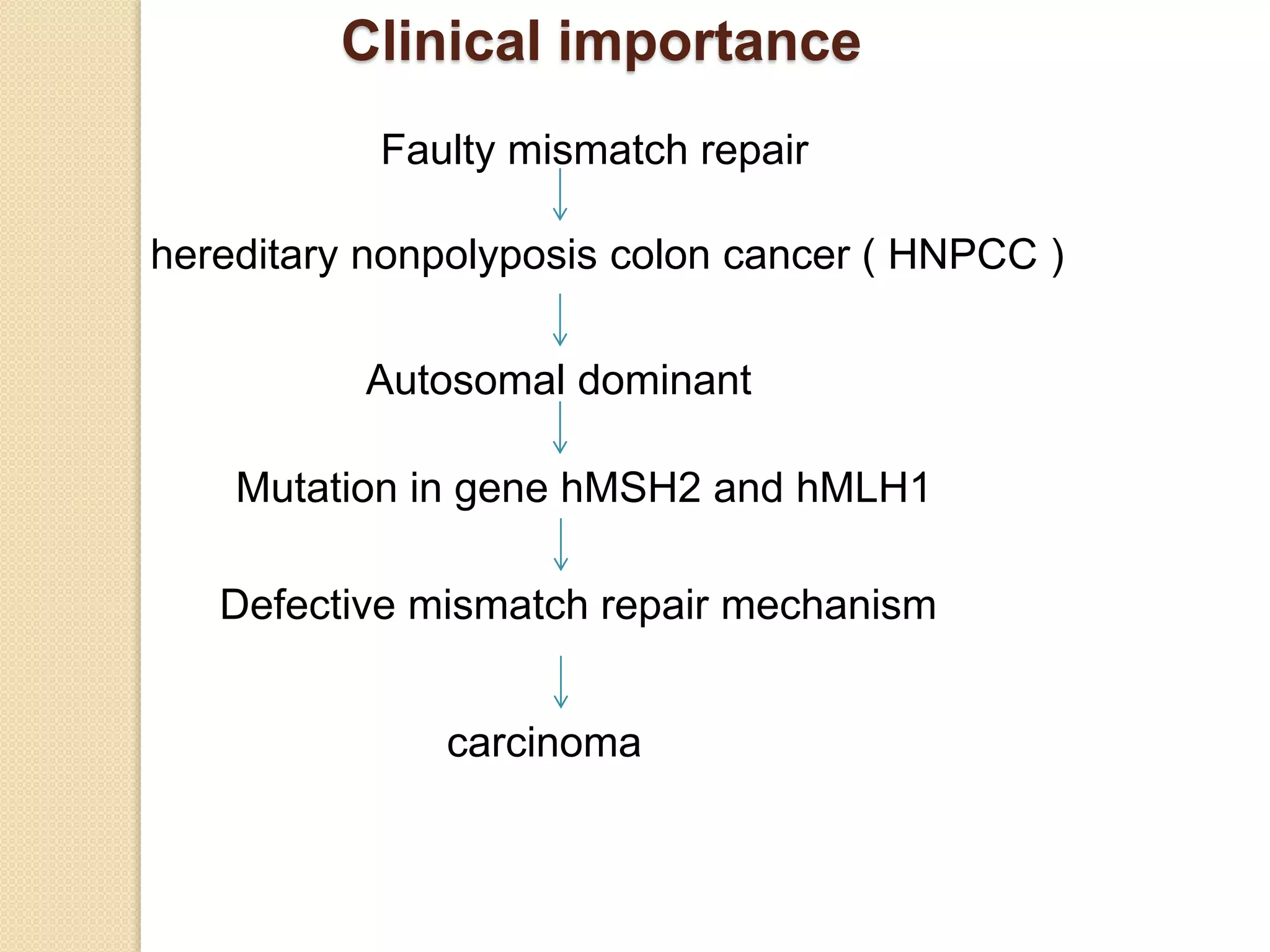
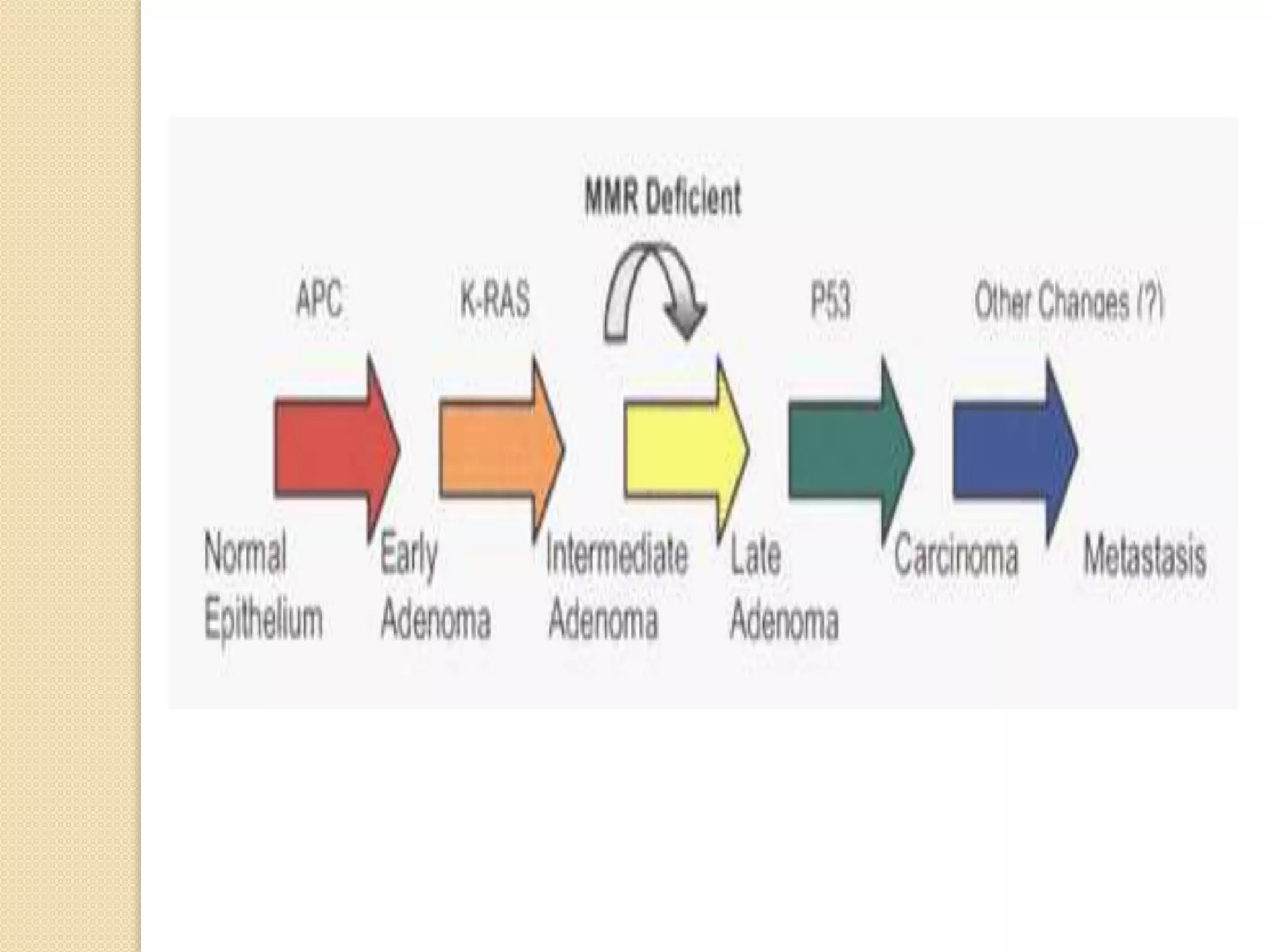
3. Defects in DNA repair pathways can lead to increased mutations, cancer, and cell death if damage is left unrepaired.