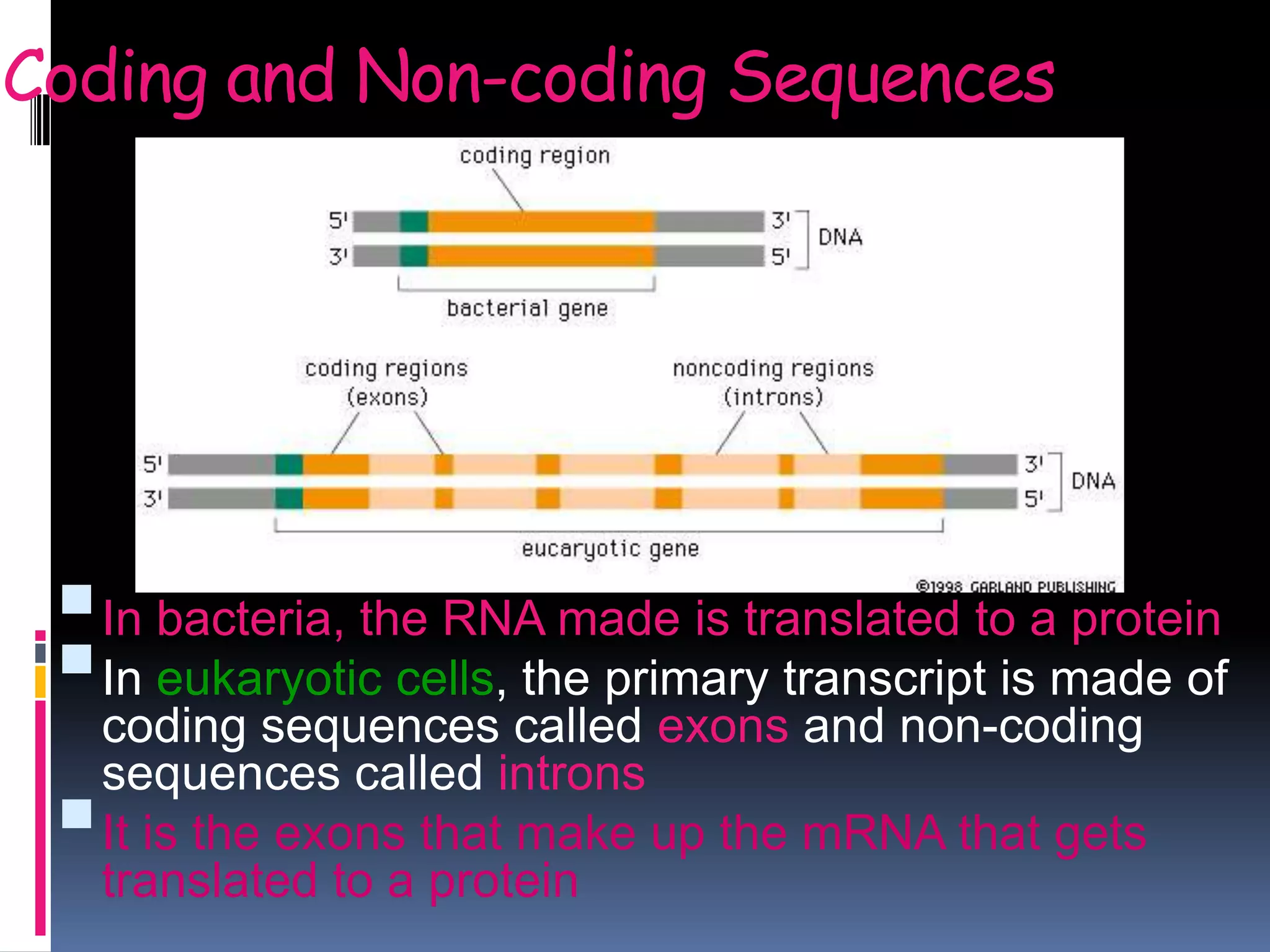
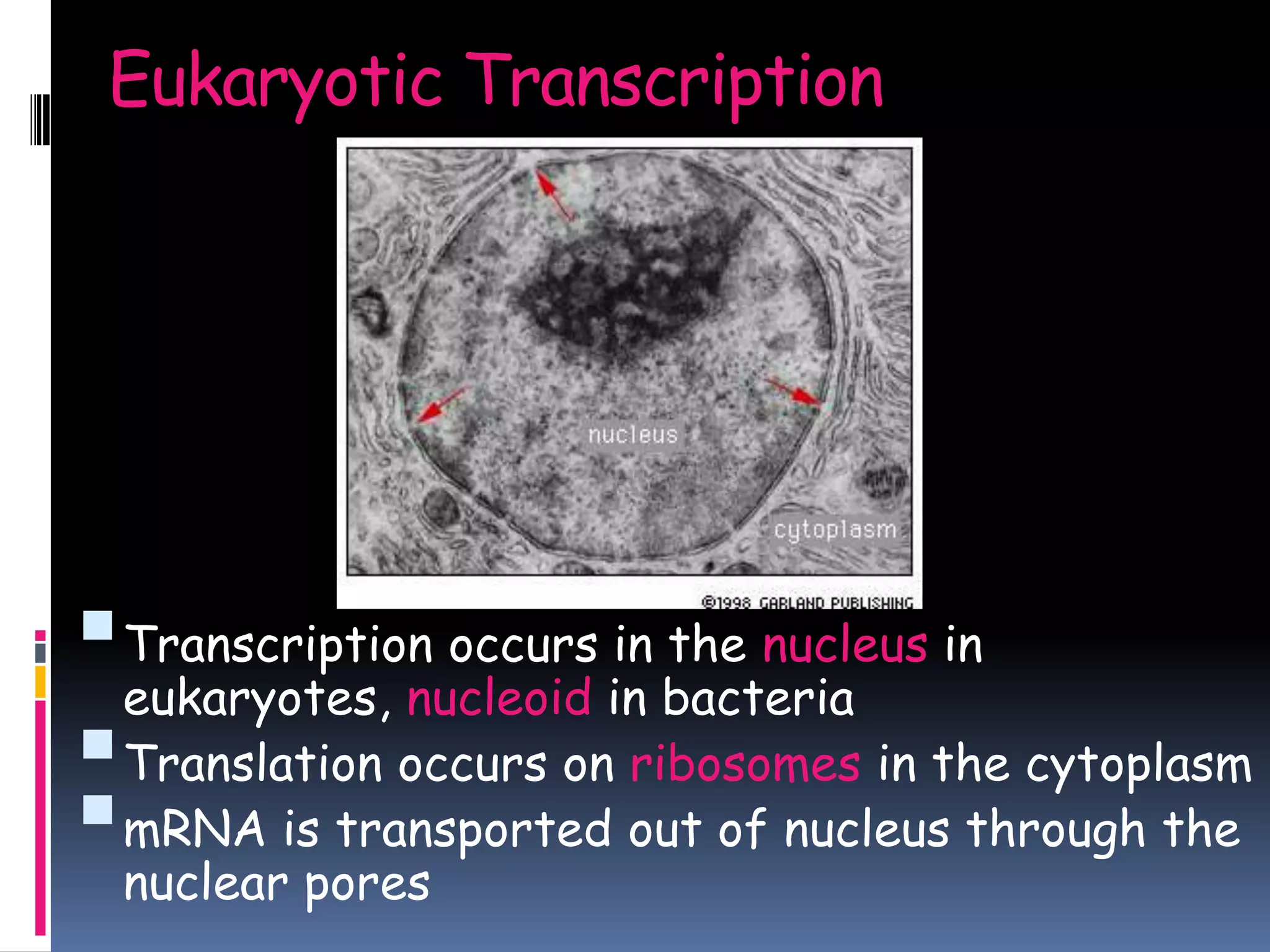
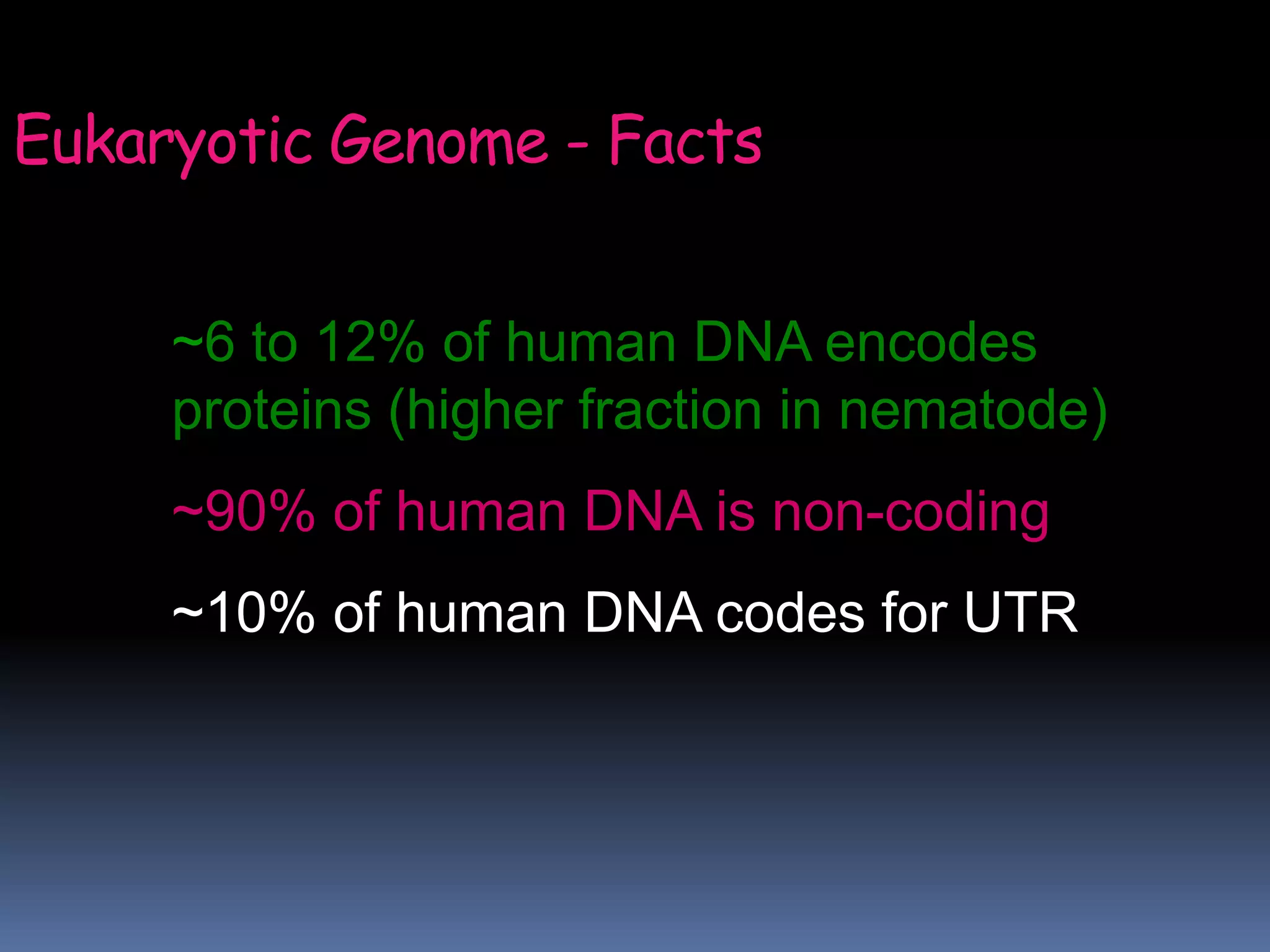
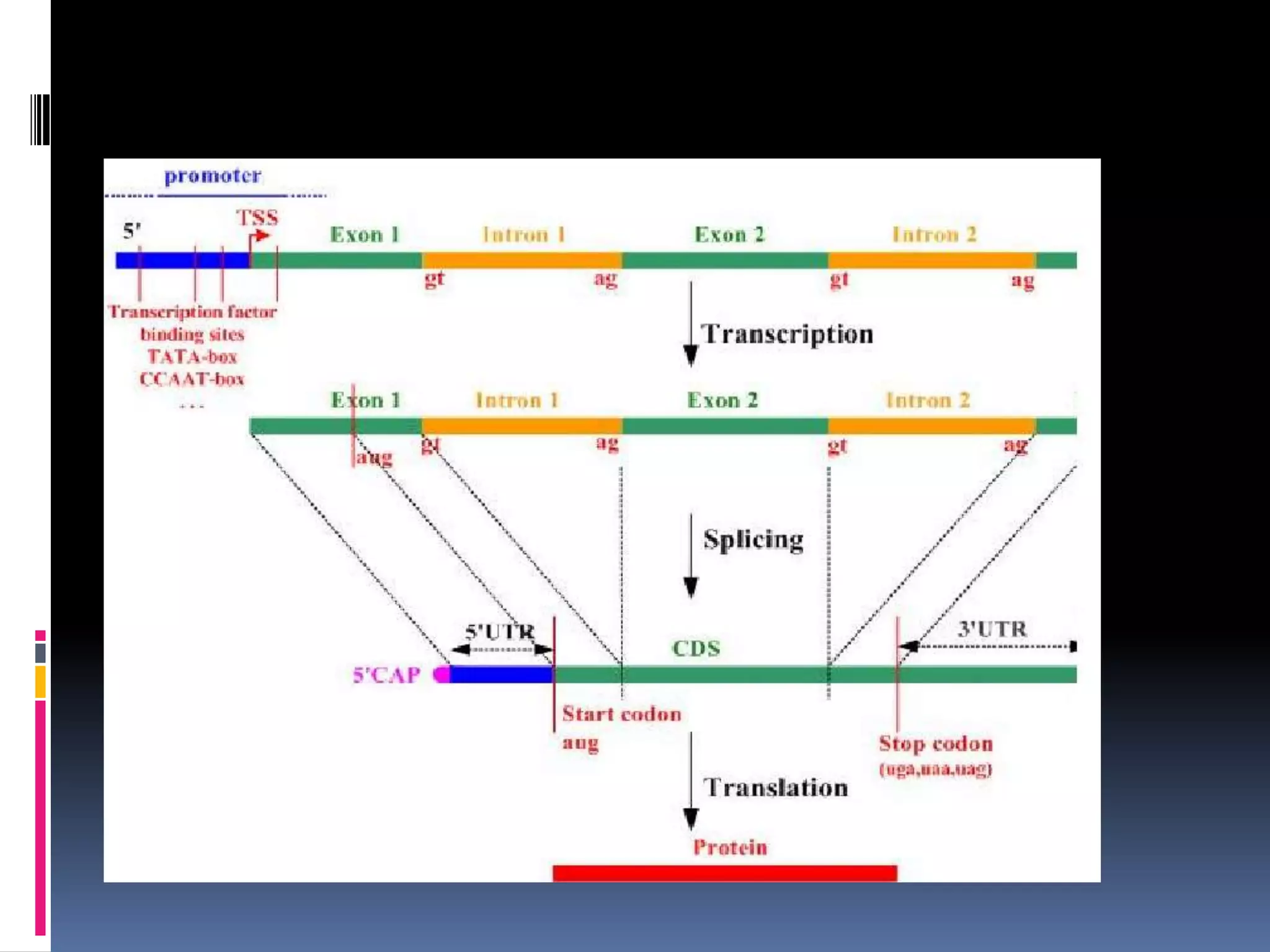
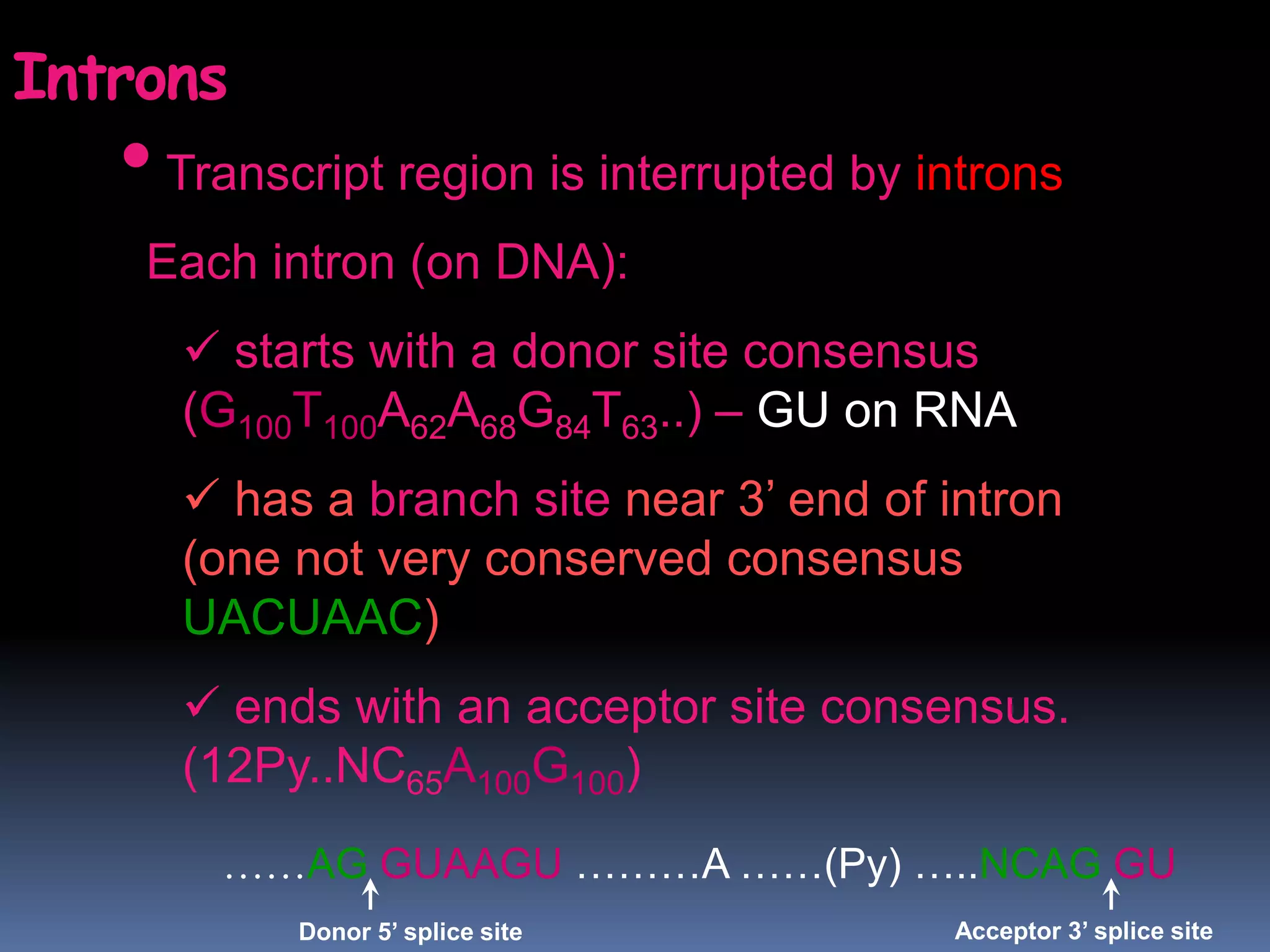
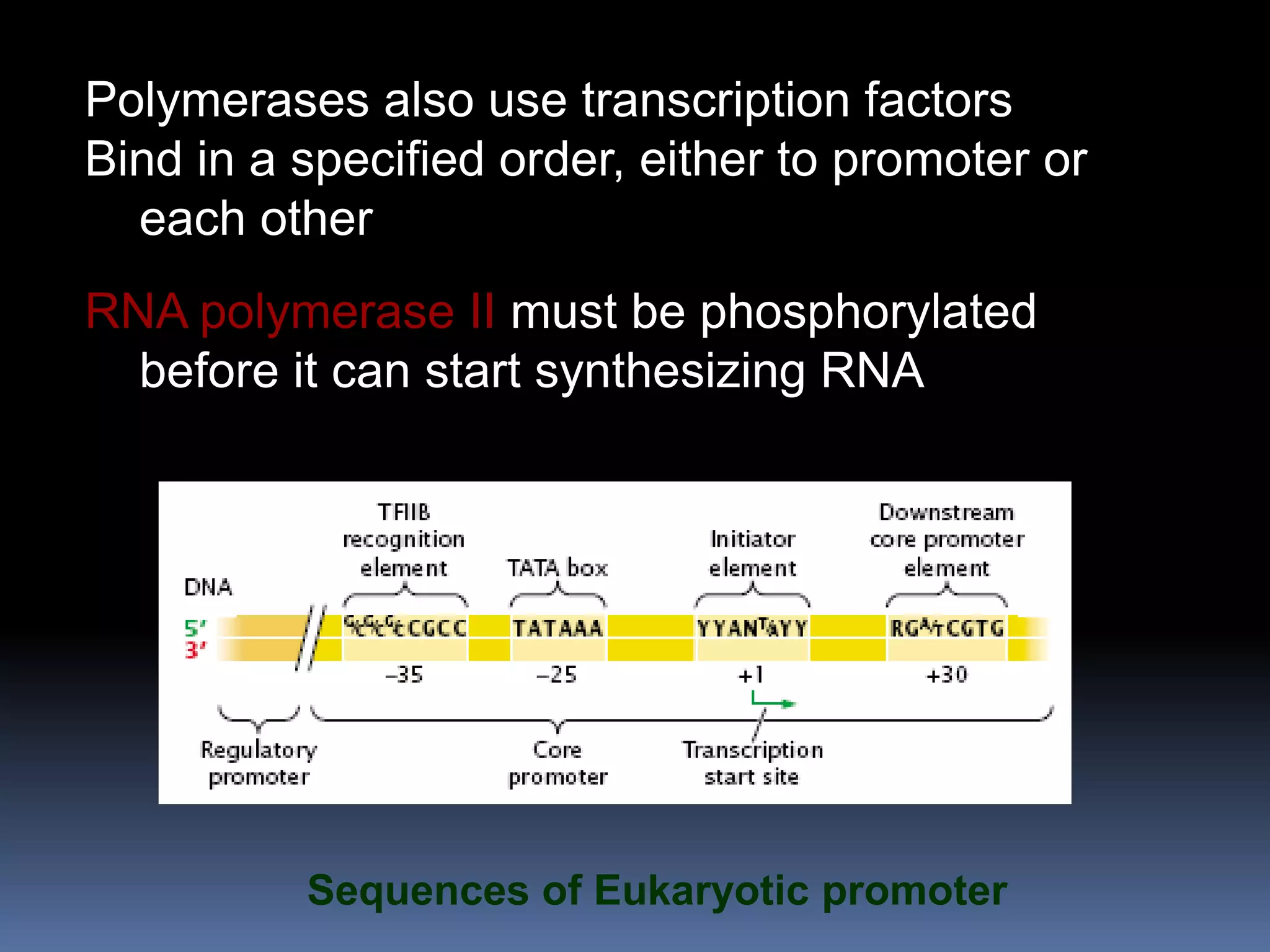
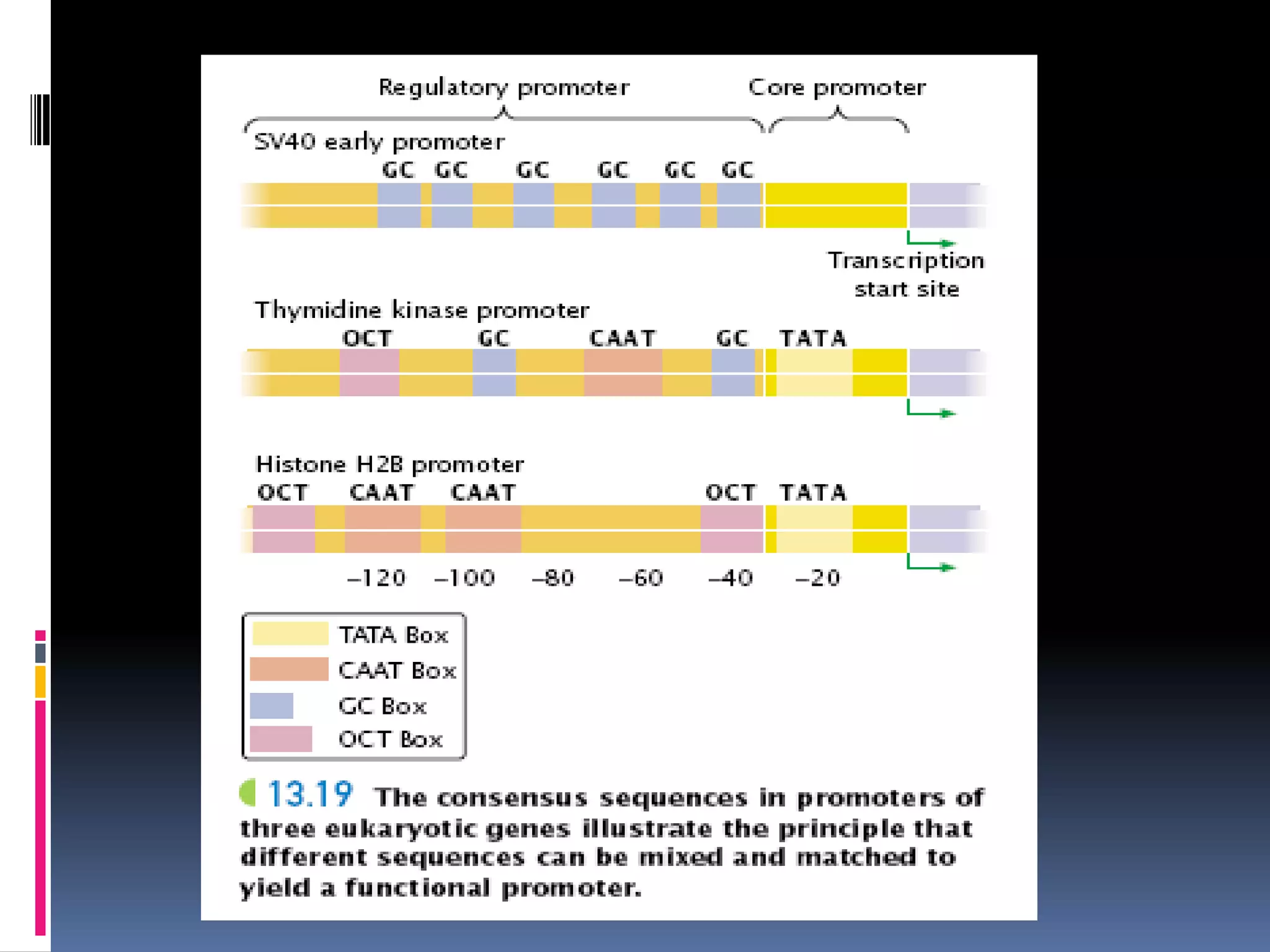
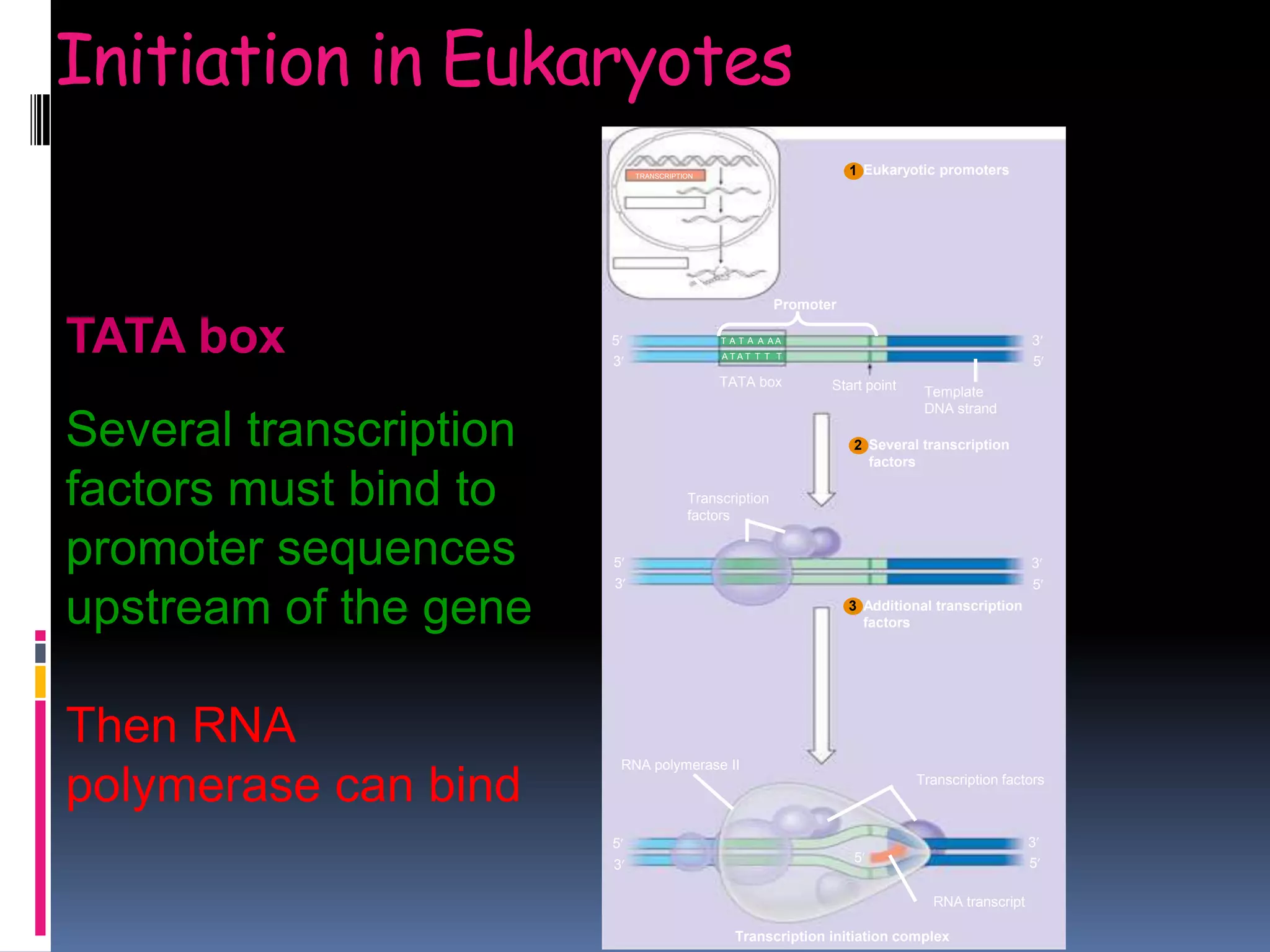
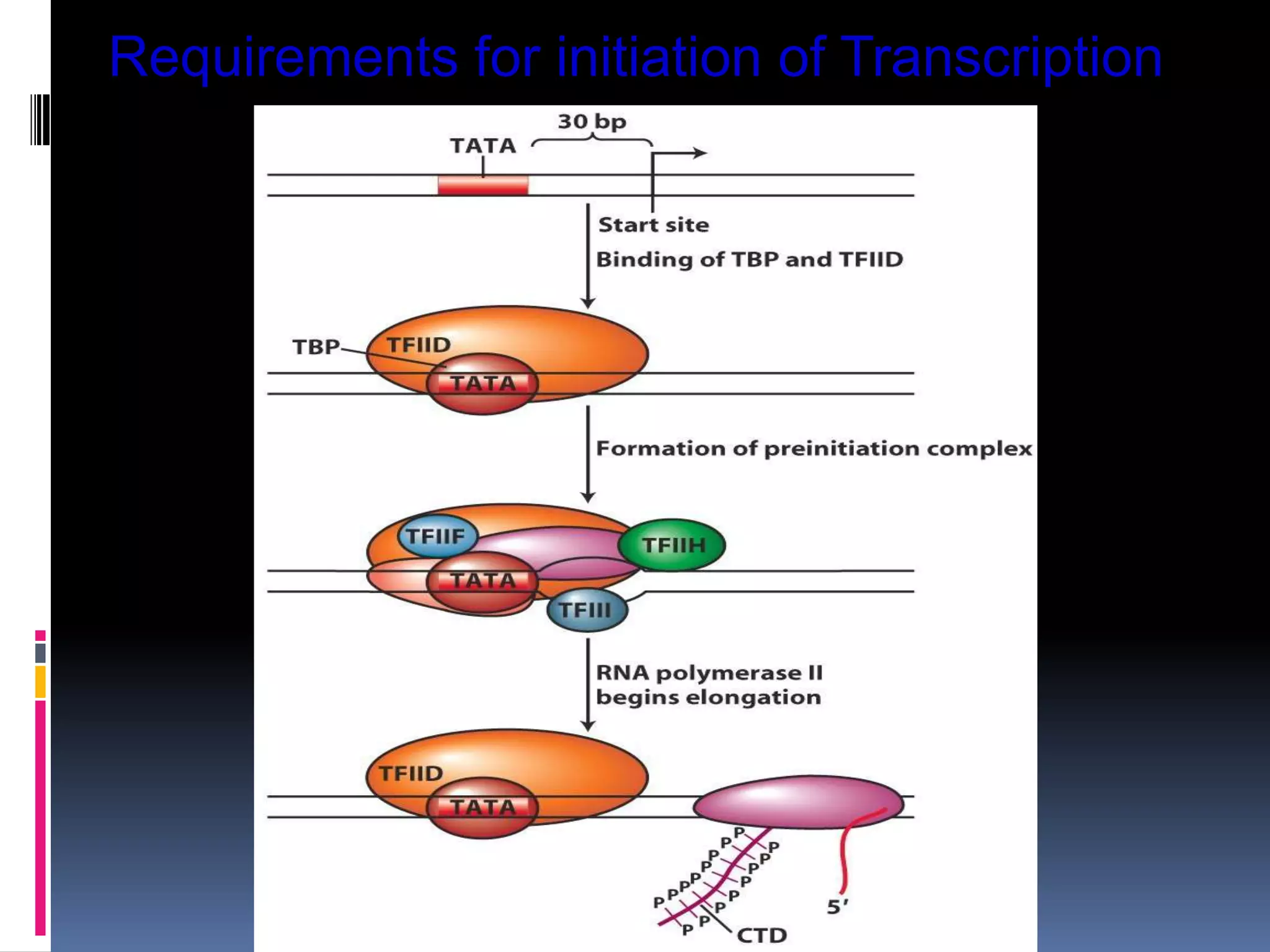
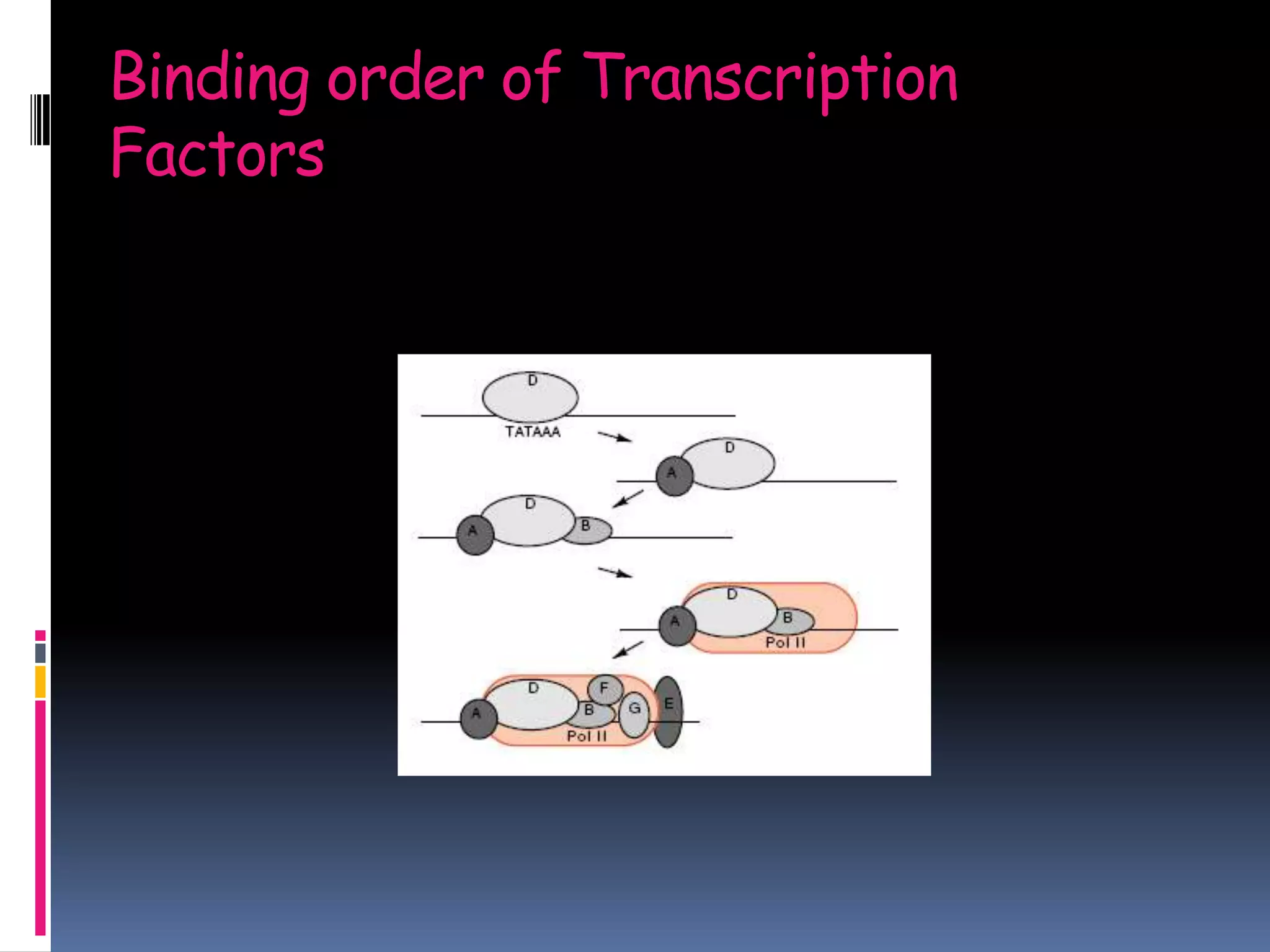
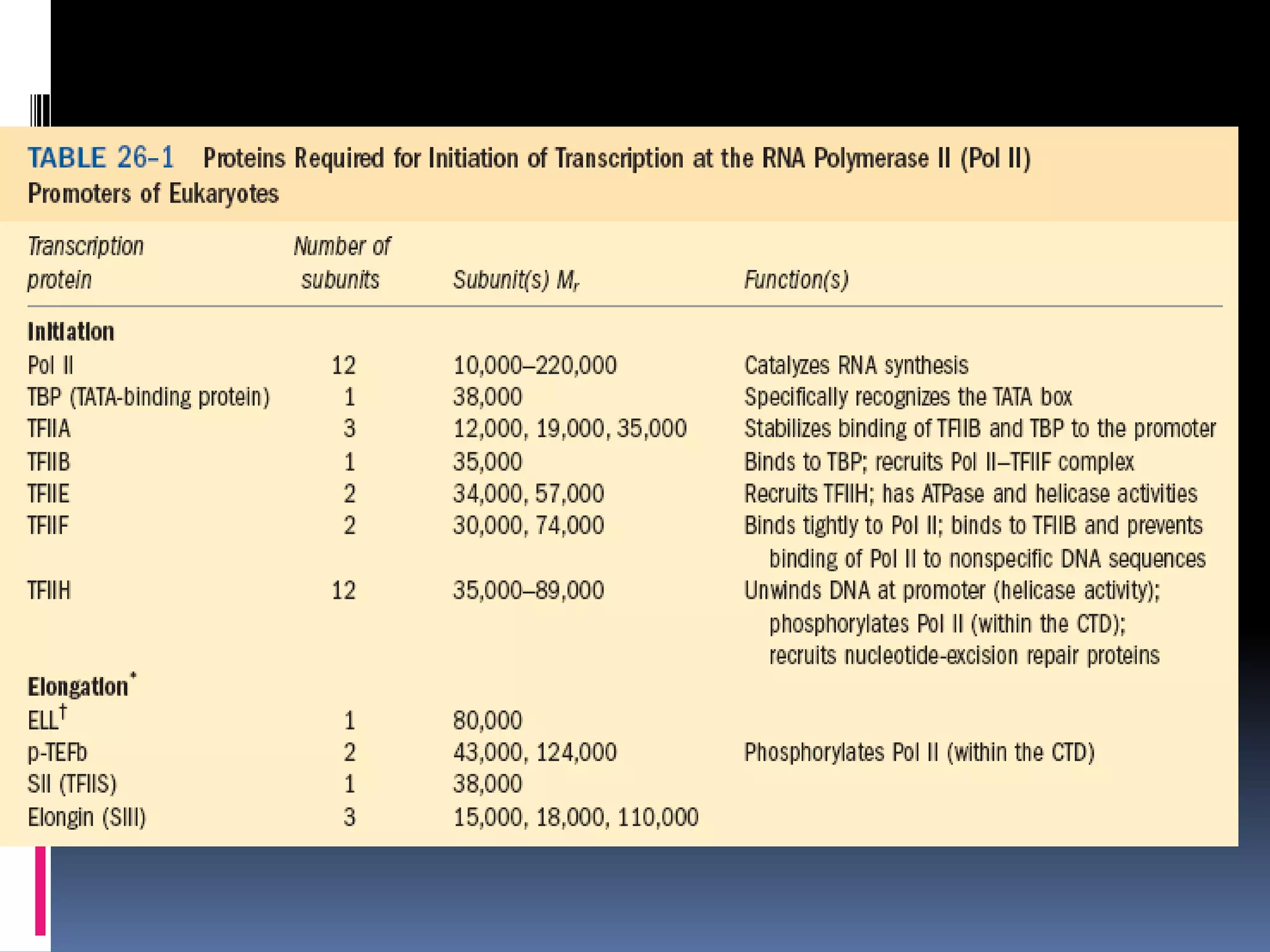
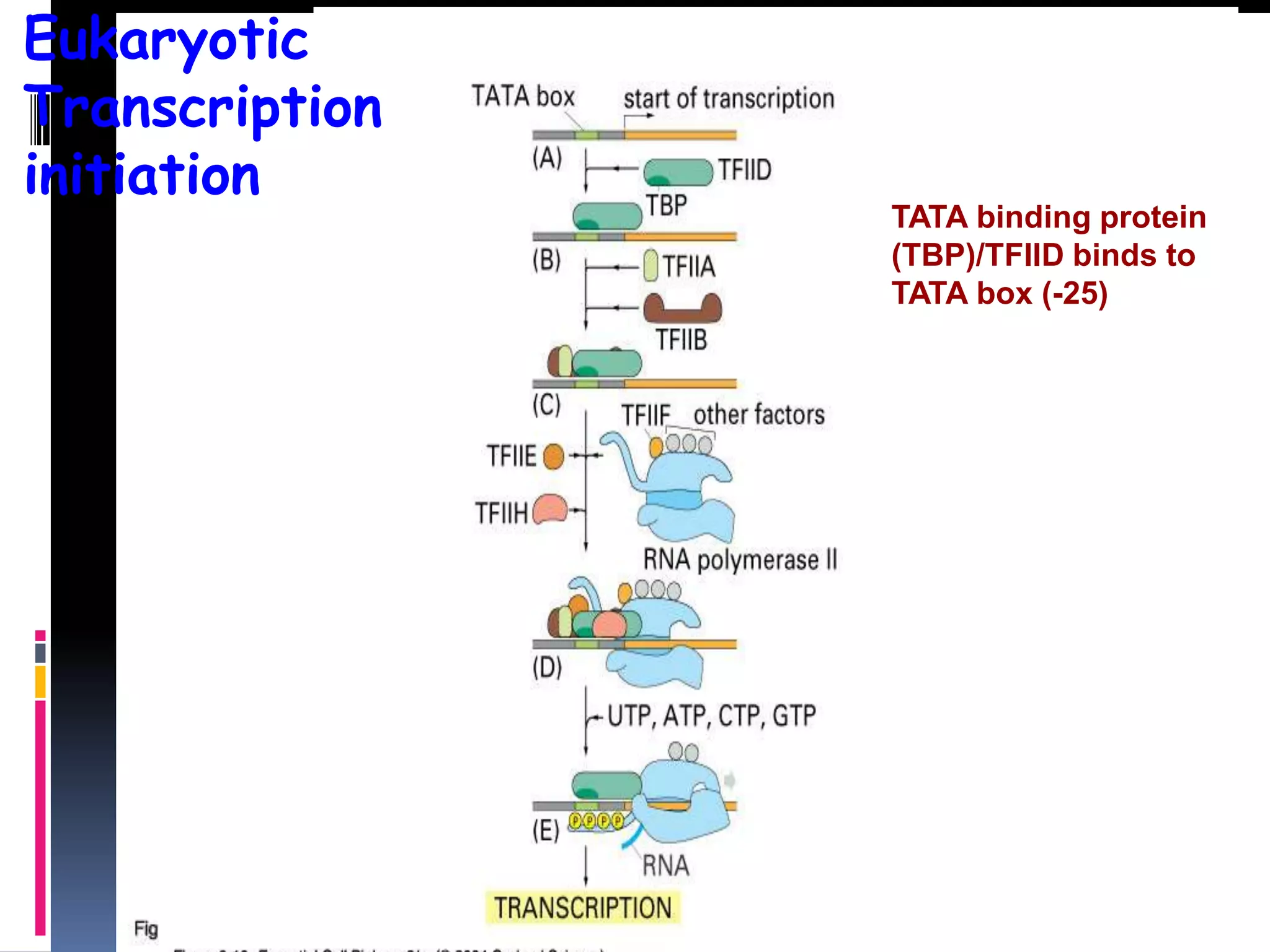
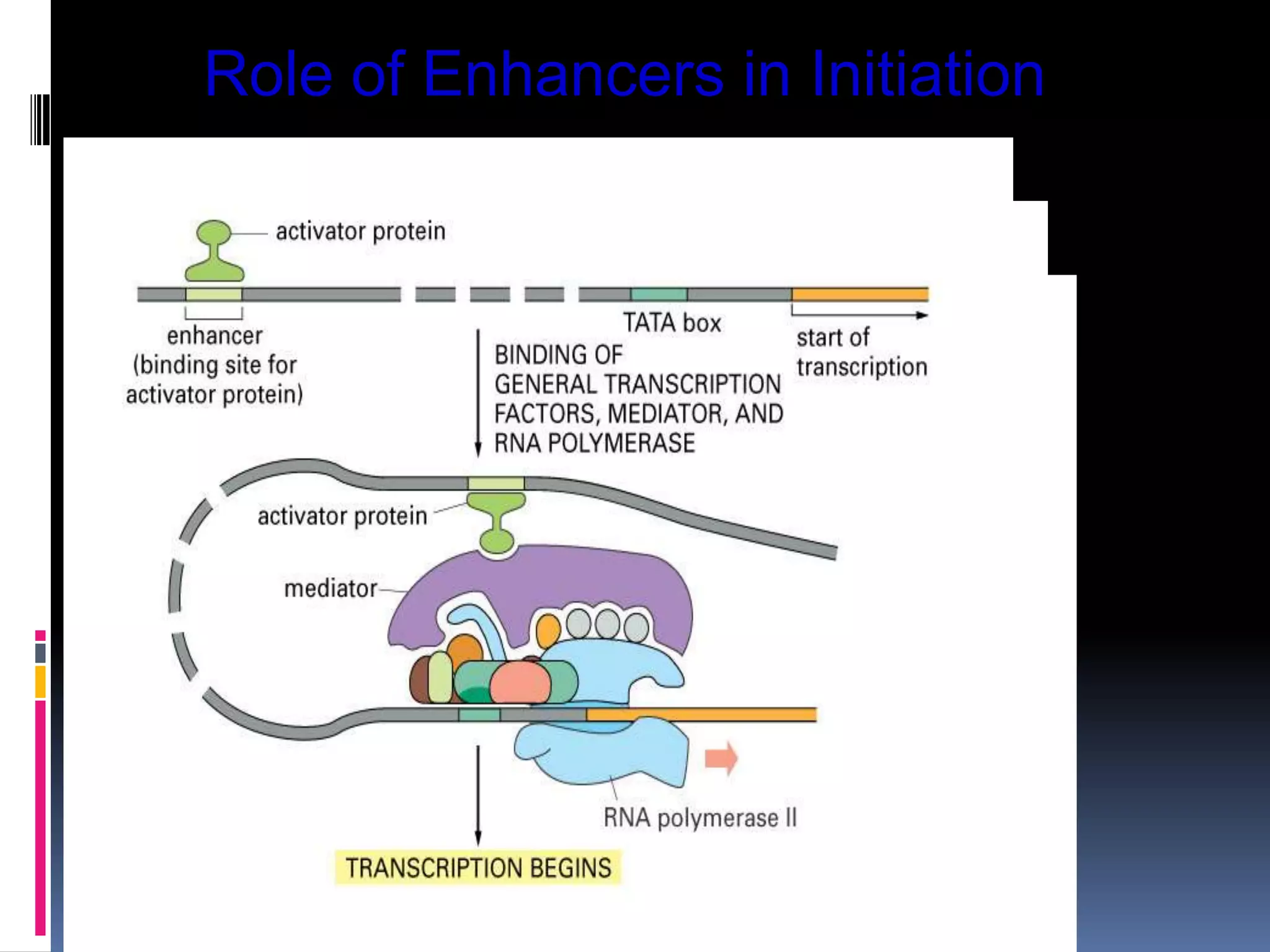
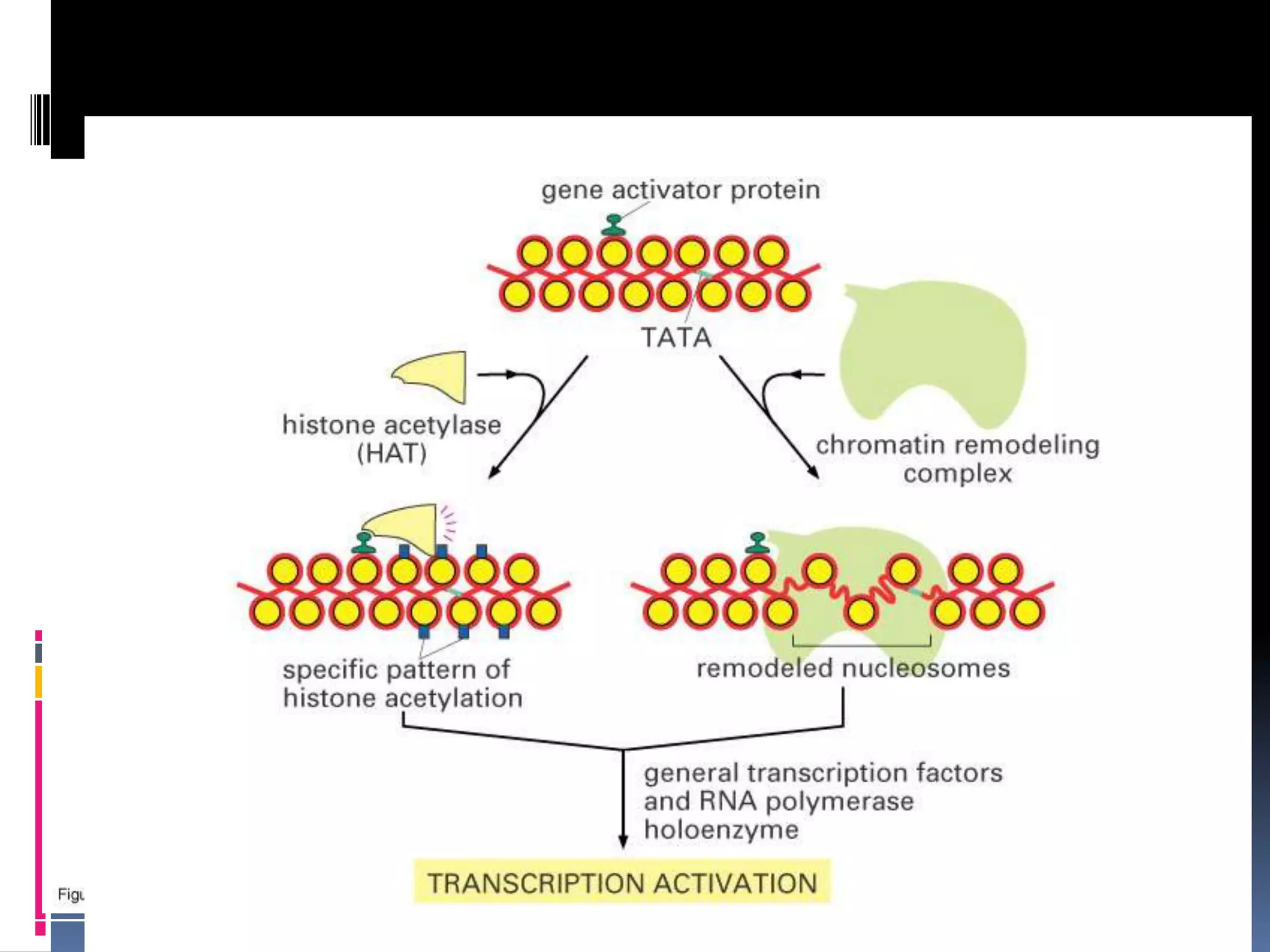
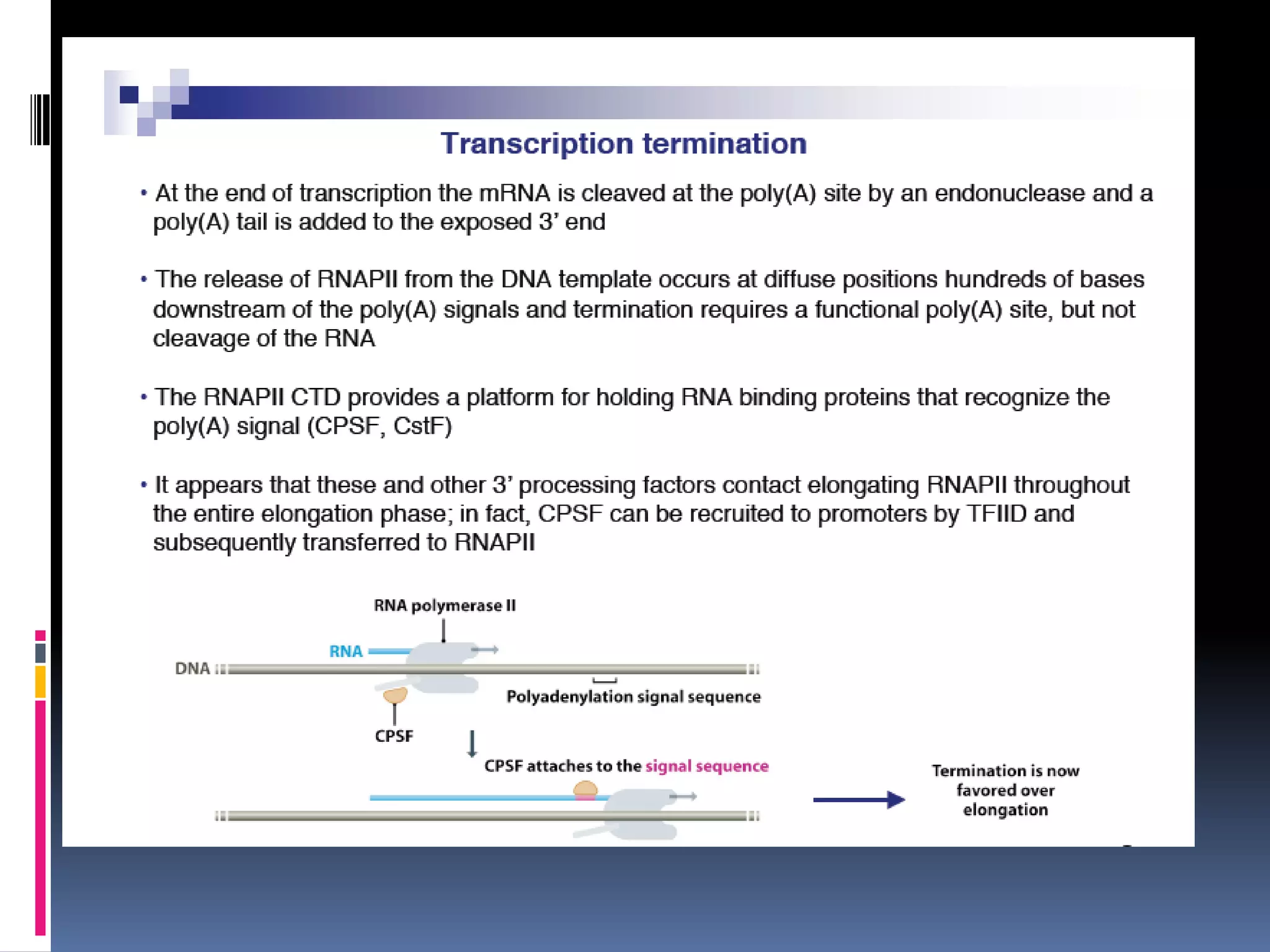
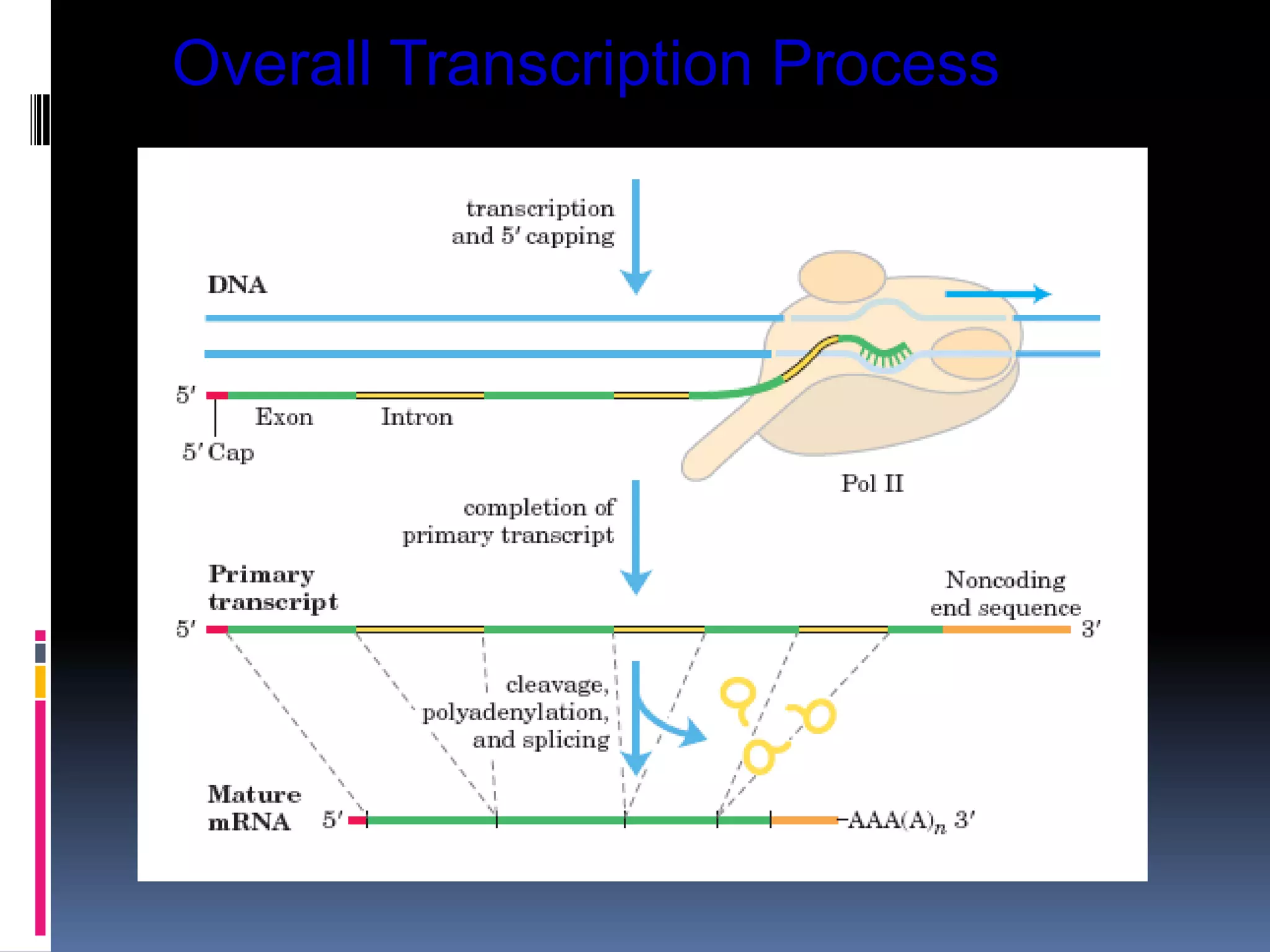
Gene expression in eukaryotes involves transcription of DNA in the nucleus, processing and export of mRNA, and translation of mRNA on ribosomes in the cytoplasm. Eukaryotic genes contain both coding exons and non-coding introns; the exons must be spliced together to form mRNA before translation. Three RNA polymerases transcribe different types of RNA in the nucleus, with RNA polymerase II responsible for mRNA and some small nuclear RNAs. Precise binding of transcription factors to promoter elements upstream of genes is required for transcription initiation by RNA polymerase II.