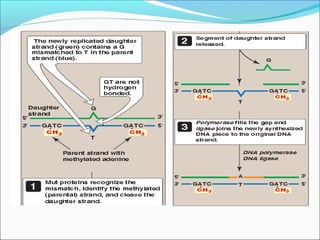
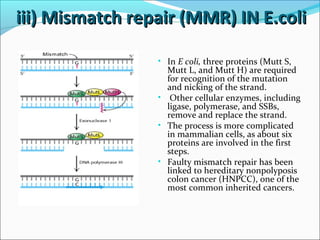
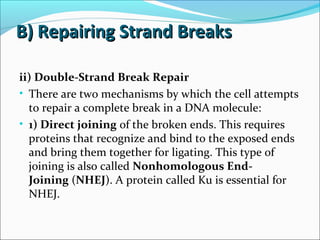
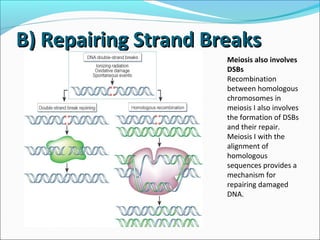
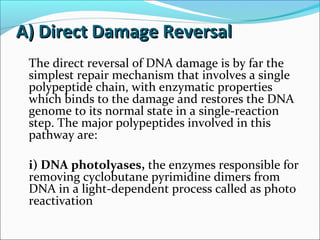
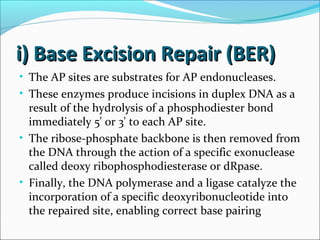
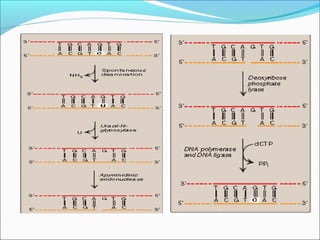
The document discusses DNA damage and repair mechanisms in human cells. It begins by explaining how errors can occur during DNA replication and how environmental agents like chemicals and radiation can damage DNA. It then describes different types of DNA damage and several pathways cells use to repair damaged DNA, including direct reversal, base excision repair, nucleotide excision repair, and mismatch repair. It provides details on these repair mechanisms and how they work to recognize and remove damaged sections of DNA before replacing them. The document concludes by discussing some genetic diseases associated with defects in DNA repair systems, like xeroderma pigmentosum, progeria, and Werner syndrome.











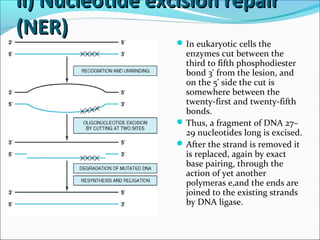
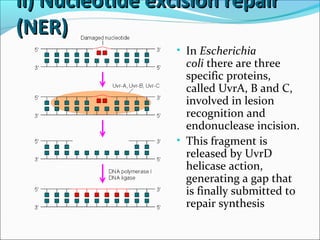
![ii) Nucleotide excision repairii) Nucleotide excision repair
(NER)(NER)
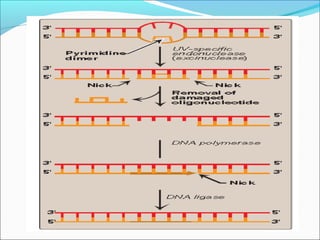
• This mechanism is used to replace regions of damaged
DNA up to 30 bases in length.
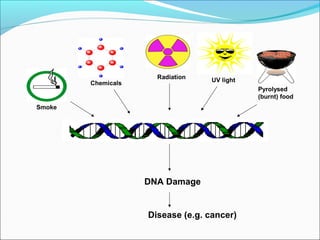
• Common causes of such DNA damage include ultraviolet
(UV) light, which induces the formation of cyclobutane
pyrimidine-pyrimidine dimers, and smoking, which causes
formation of benzo[a]pyrene-guanine adducts.
• Ionizing radiation, cancer chemotherapeutic agents, and a
variety of chemicals found in the environment cause base
modification, strand breaks, cross-linkage between bases
on opposite strands or between DNA and protein, and
numerous other defects.
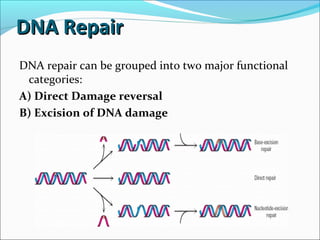
• These are repaired by a process called nucleotide excision-
repair](https://image.slidesharecdn.com/dnadamage-190216131554/85/Dna-damage-lgis-19-320.jpg)