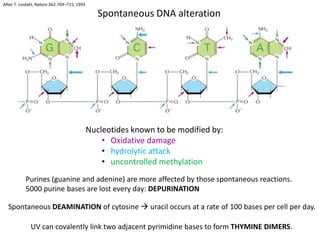
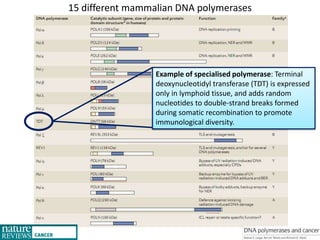
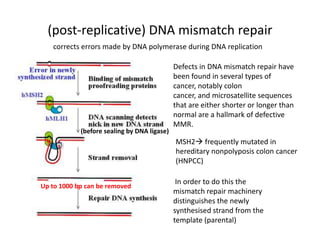
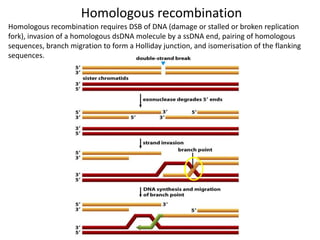
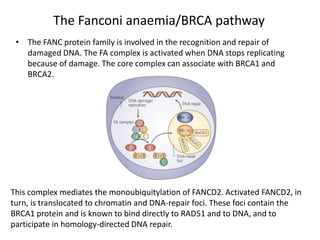
Genetic stability relies on accurate DNA replication and repair mechanisms to correct approximately 1 million lesions per cell per day caused by various sources of DNA damage. If unrepaired, this damage can lead to mutations, genetic diseases like cancer, or cell death. The cell has multiple pathways to repair DNA damage, including base excision repair, nucleotide excision repair, mismatch repair, and homologous recombination. Homologous recombination is especially important for repairing double-stranded breaks using the undamaged sister chromatid as a template, and involves proteins such as RAD51, BRCA1, BRCA2, ATM, and the Fanconi anemia/BRCA pathway. Defects in DNA repair genes are