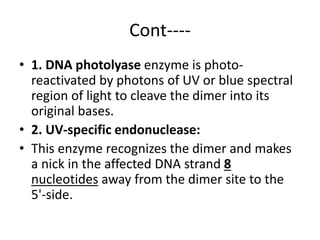
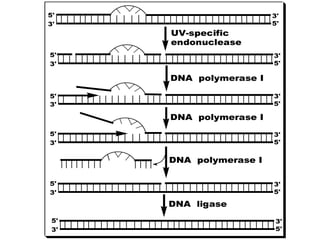
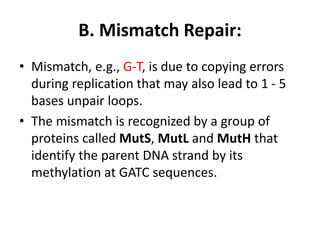
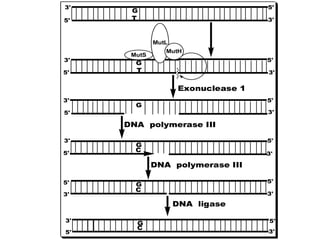
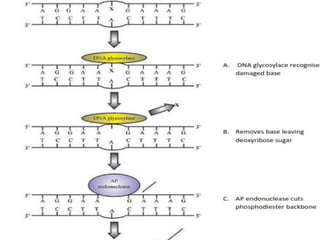
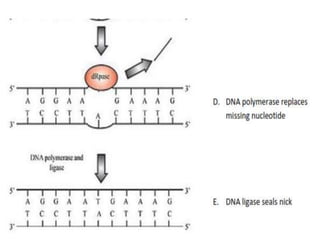
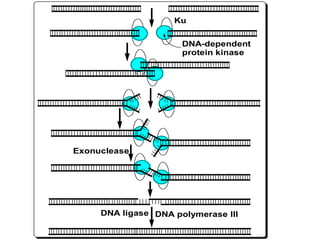
DNA mutations can occur through various causes like radiation, chemicals, and replication errors. There are several systems that repair DNA damage to prevent mutations:
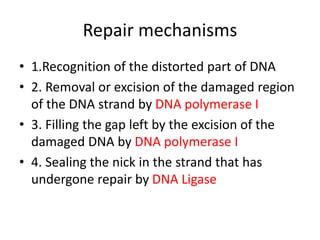
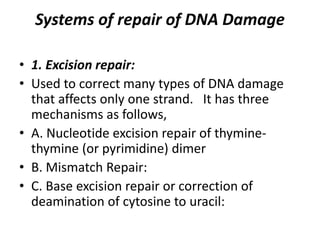
1. Excision repair removes and replaces damaged DNA sections through mechanisms like nucleotide excision repair of thymine dimers, mismatch repair of incorrect bases, and base excision repair of deaminated bases.
2. Recombinational repair is used for double-strand breaks, where proteins like Ku and DNA-dependent protein kinase recognize the breaks and use complementary DNA sequences to reconnect the strands. Any gaps are then filled in and sealed.