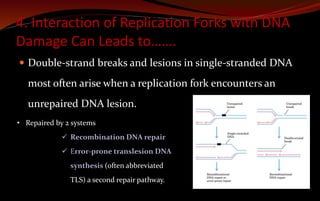
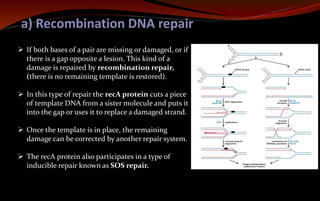
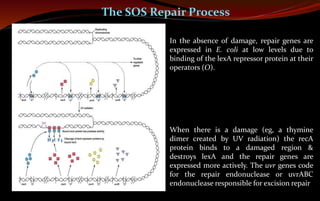
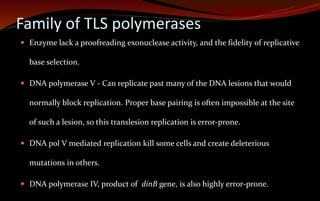
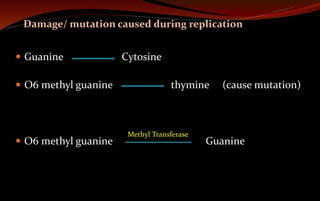
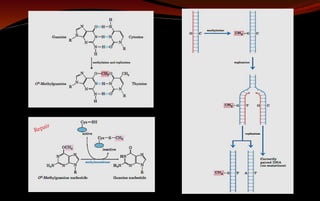
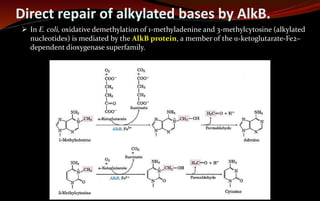
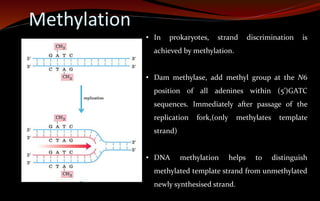
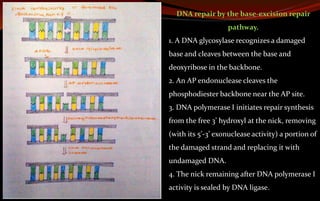
The document discusses various DNA repair mechanisms addressing how DNA can be damaged by environmental factors and errors during replication. It details multiple repair systems, including direct repair, mismatch repair, base-excision repair, and nucleotide-excision repair, explaining their processes and the enzymes involved. Additionally, it highlights the importance of these repair pathways in maintaining genomic integrity and their links to human diseases when defective.









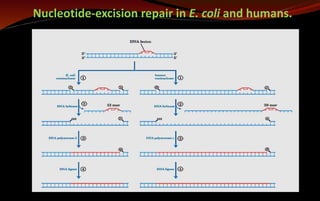
![ In E. coli and other prokaryotes – it generates 12 to 13 nucleotides fragment.
In humans and other eukaryotes - produce 27 to 29 nucleotides fragment.
The excised oligonucleotides are released from the duplex and the resulting gap is
filled—by DNA polymerase I in E. coli and DNA polymerase ε in humans. DNA ligase
seals the nick.
This pathway is a primary repair route for many types of lesions, including
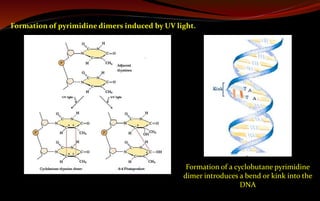
cyclobutane pyrimidine dimers, 6-4 photoproducts
other types of base adducts including benzo[a]pyrene-guanine, which is formed in
DNA by exposure to cigarette smoke.
Nucleotide-excision repair and base-excision repair in eukaryotes is closely tied to
transcription.](https://image.slidesharecdn.com/dnarepair-240123020228-d0ecf534/85/DNA-Repair-Mechanisms-pptx-22-320.jpg)