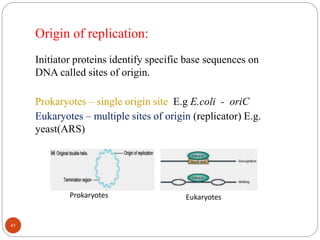
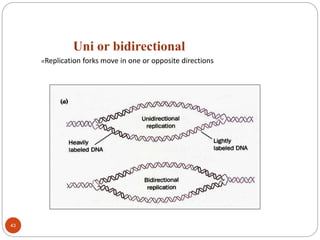
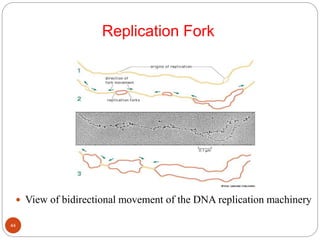
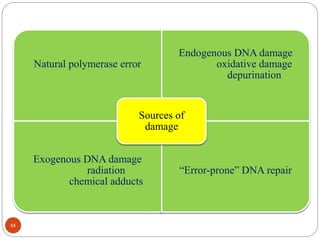
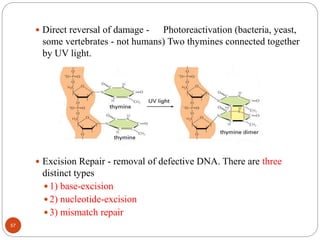
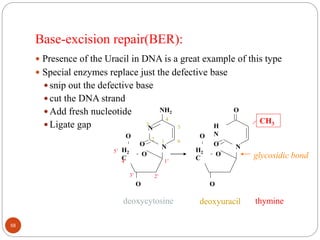
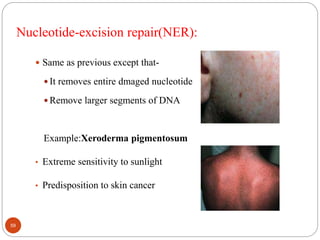
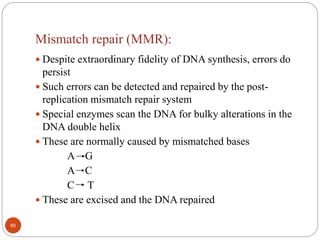
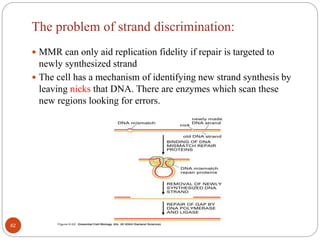
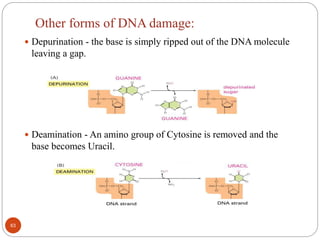
The document provides information on DNA, including its identification as the genetic material, its structure and replication. Some key points:
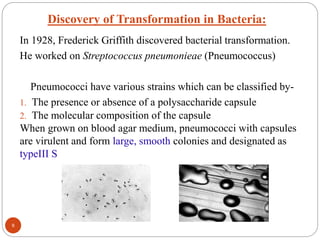
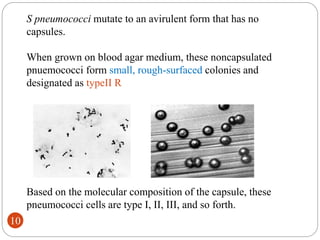
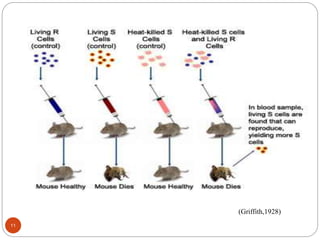
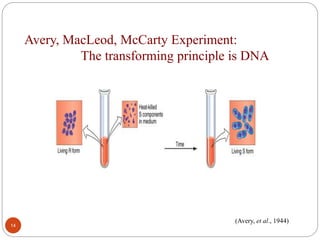
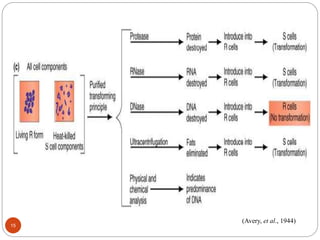
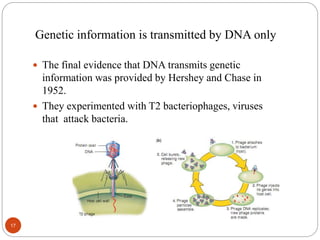
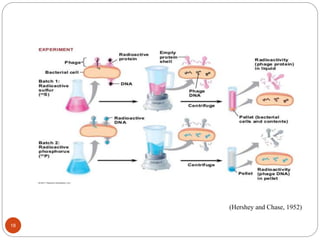
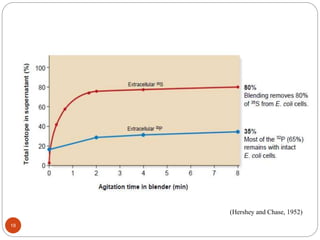
- Experiments in the 1920s-1950s identified DNA as the genetic material, including Griffith's work on bacterial transformation and the experiments of Avery, MacLeod and McCarty and Hershey and Chase.
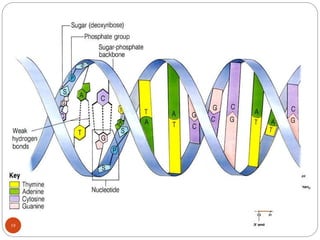
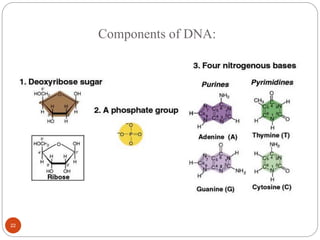
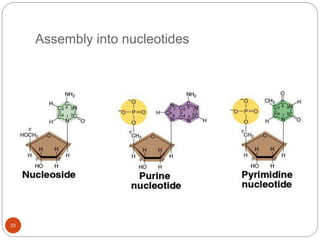
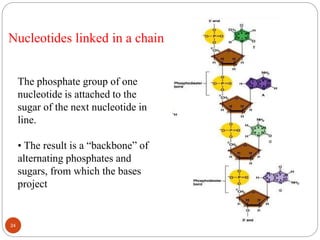
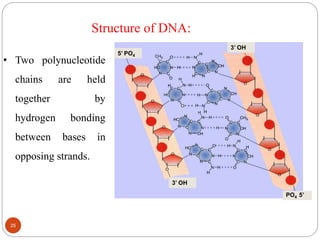
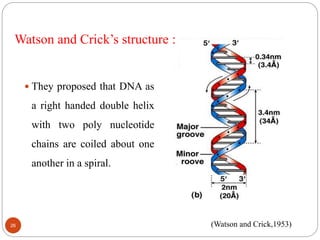
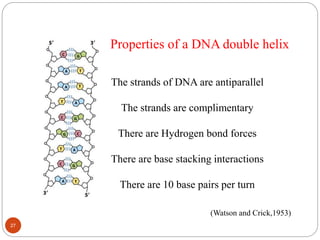
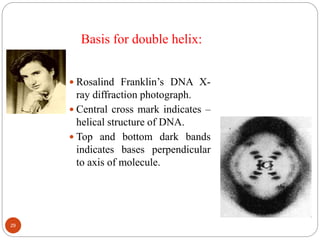
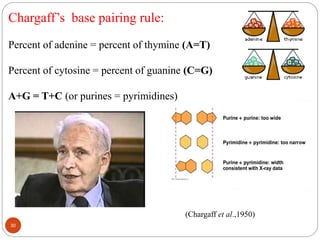
- DNA is made up of nucleotides containing phosphate groups, sugars and nitrogenous bases. Watson and Crick discovered its double helix structure in 1953, with two anti-parallel strands held together by hydrogen bonds between complementary bases.
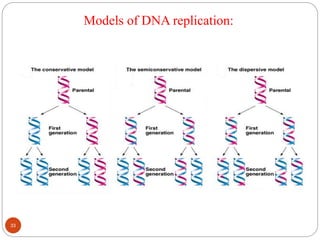
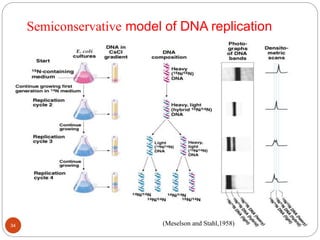
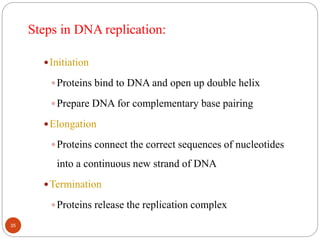
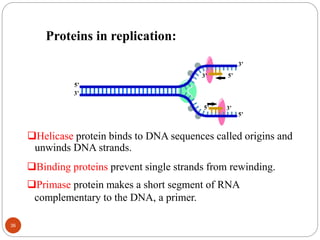
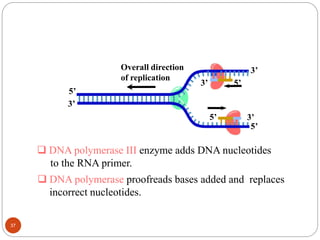
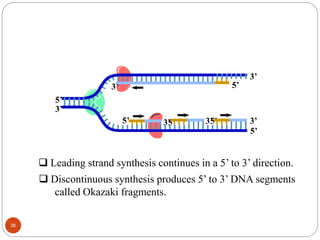
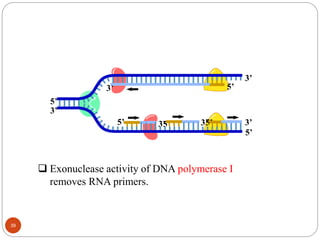
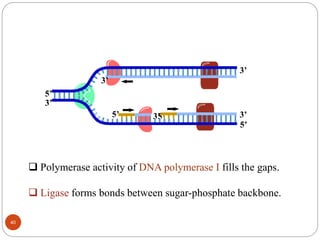
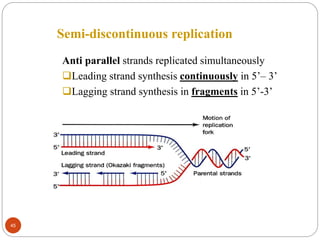
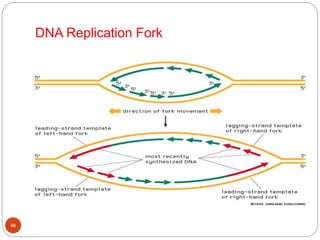
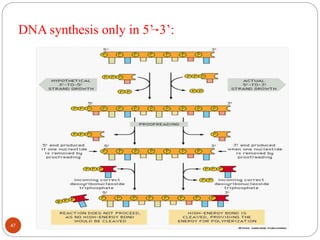
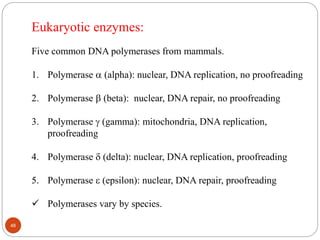
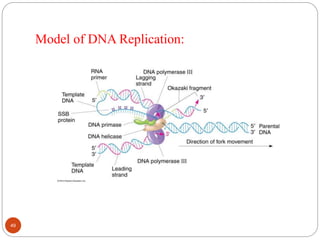
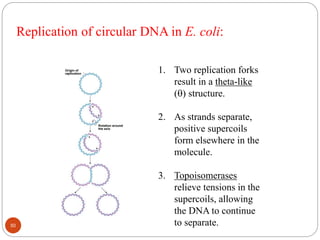
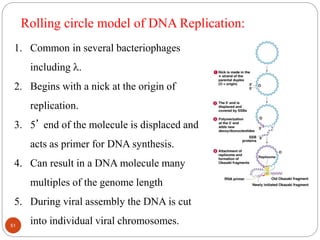
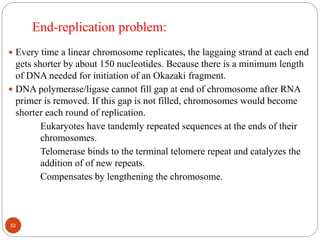
- DNA replication is semi-conservative and involves unwinding of the DNA strands, synthesis of new complementary strands, and production of identical double