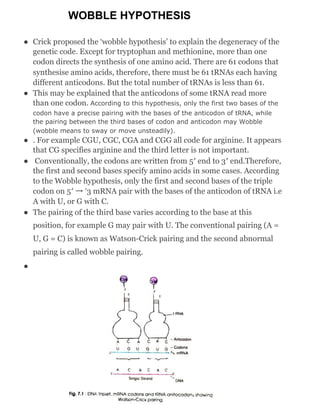
The wobble hypothesis, proposed by Crick, explains how some tRNA can recognize multiple codons for the same amino acid due to flexible pairing at the third codon position. This hypothesis asserts that only the first two bases of a codon pair precisely with a tRNA anticodon while the third can 'wobble', allowing for alternative base pairing. Wobble base pairs include combinations like guanine-uracil and hypoxanthine with various nucleotides, enabling a single tRNA to recognize several codons.