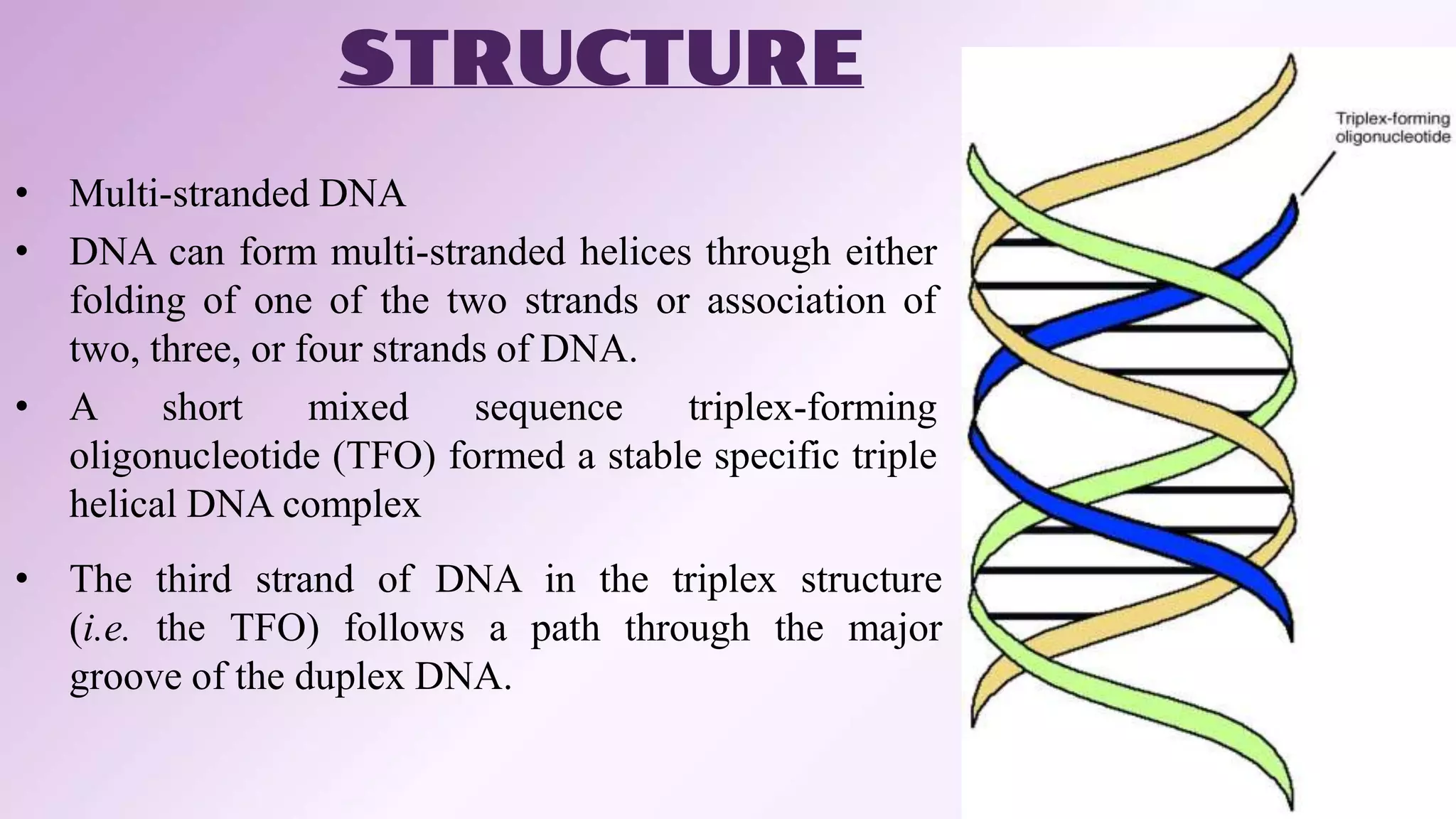
This document summarizes information about triple helix DNA structure and its potential applications. It discusses the discovery of DNA and triple helical structures. There are two classifications of triple helices: intermolecular, formed by a third strand from another DNA molecule, and intramolecular or H-DNA, formed by one strand of the same duplex. Factors like length, base composition, and temperature affect triplex formation. Potential therapeutic applications include targeting genes to modulate expression, directing site-specific DNA damage, and stimulating DNA repair. However, limitations include stability in cells and accessibility due to chromatin structure.