This document provides an overview of DNA sequencing:
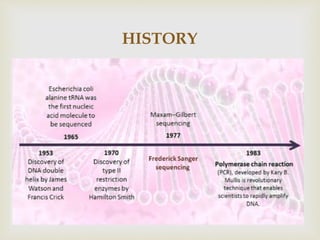
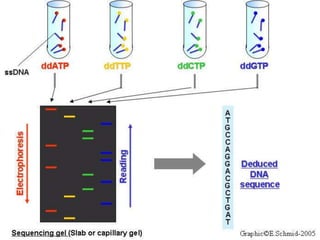
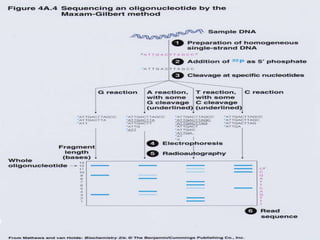
- It discusses the history of DNA sequencing, from the early 1970s methods to modern techniques. The Sanger and Maxam-Gilbert methods were among the first developed.
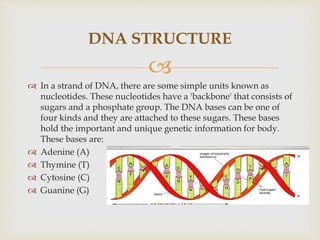
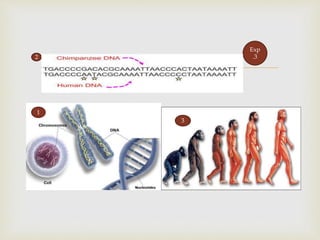
- DNA sequencing involves determining the order of nucleotides (A, T, C, G) in a DNA molecule. This provides important information for research and applications in diagnostics, biotechnology, forensics, and more.
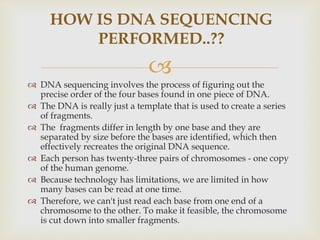
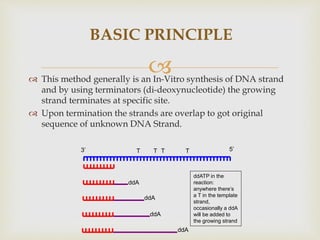
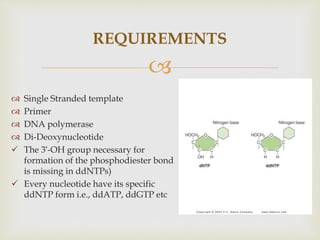
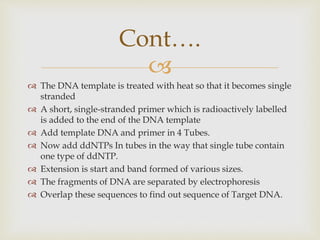
- The document outlines some of the major DNA sequencing techniques and methods, including Sanger sequencing and Maxam-Gilbert sequencing. It also discusses next-generation sequencing approaches.