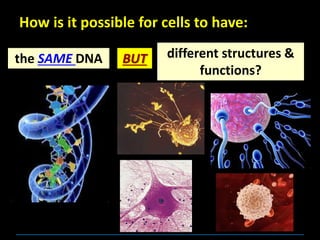
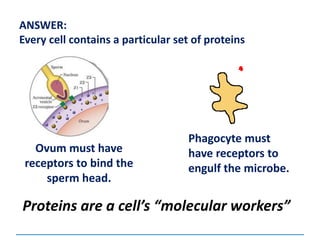
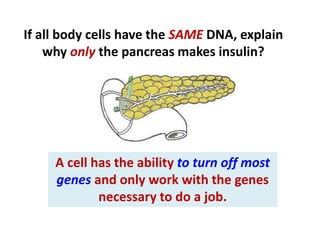
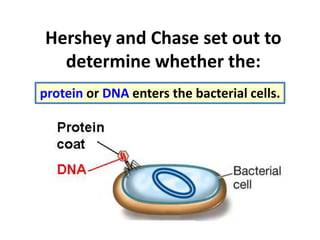
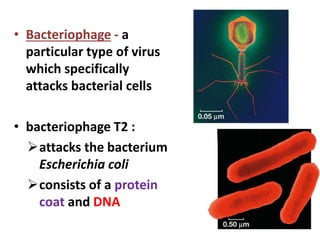
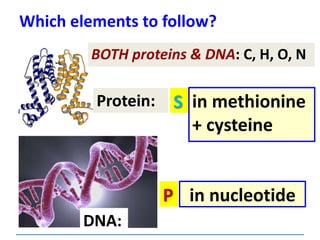
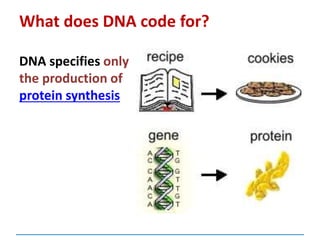
This document discusses several key aspects of DNA structure and function:
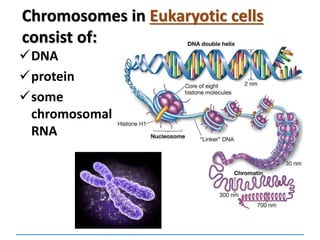
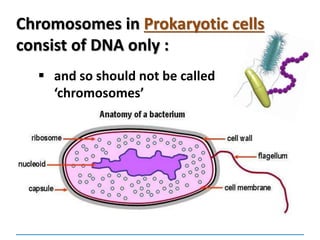
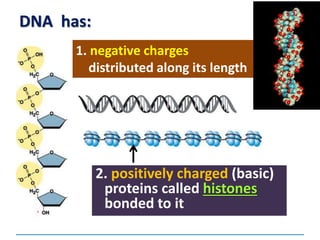
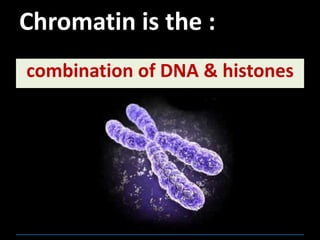
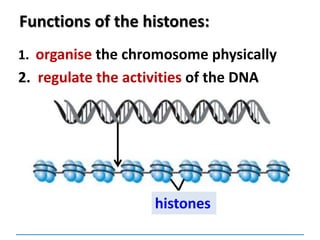
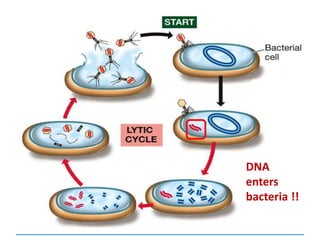
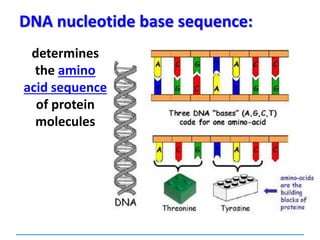
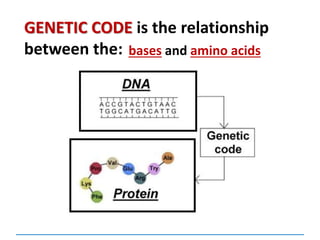
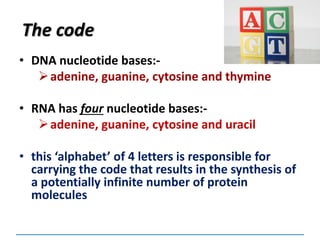
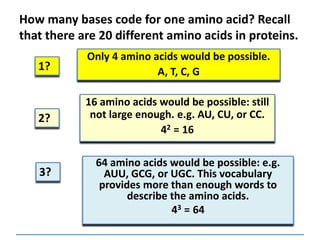
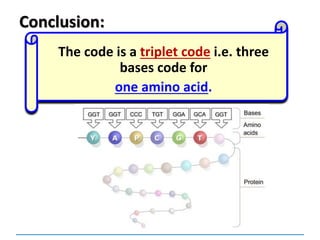
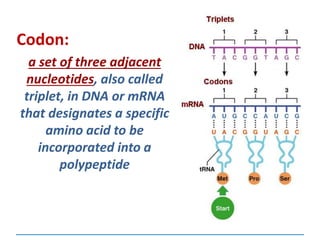
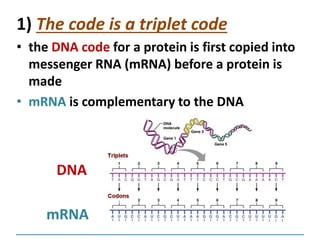
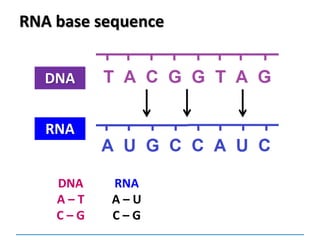
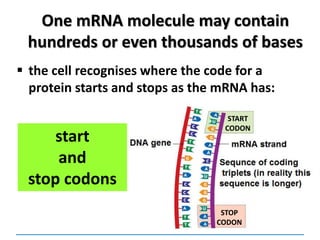
1) Chromosomes contain DNA, histone proteins, and some RNA. DNA contains a code made up of four nucleotide bases that determines the sequence of amino acids in proteins.
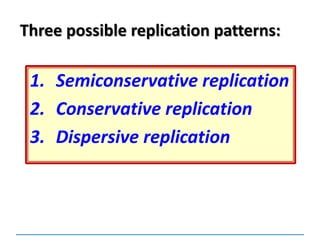
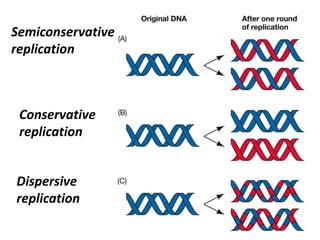
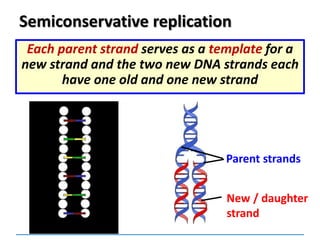
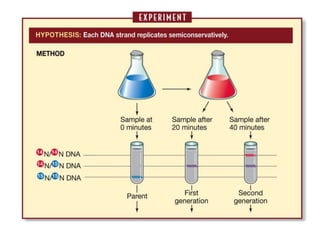
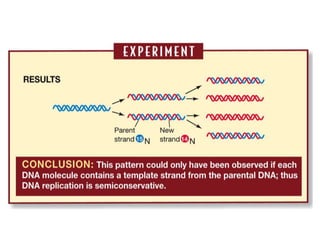
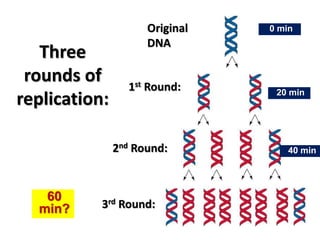
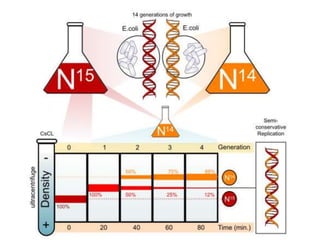
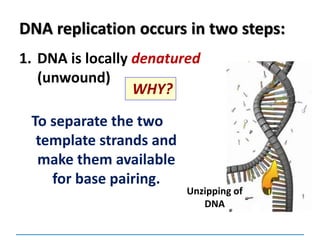
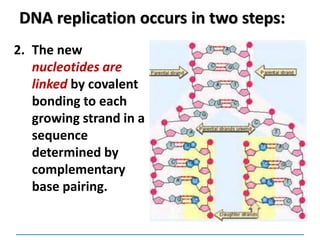
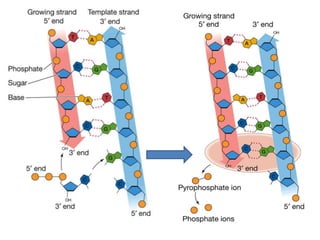
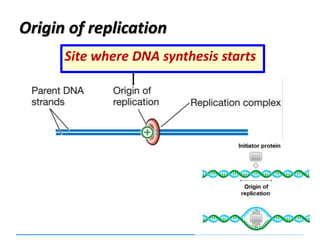
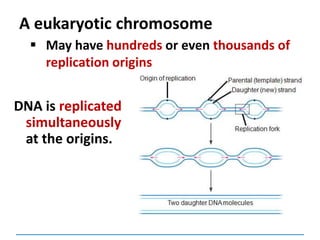
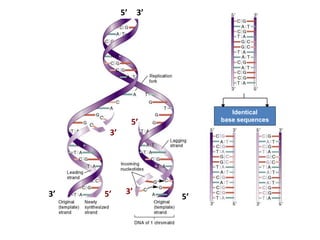
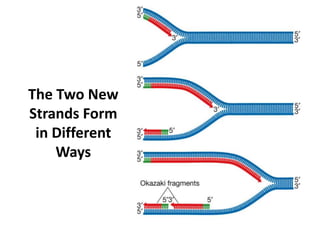
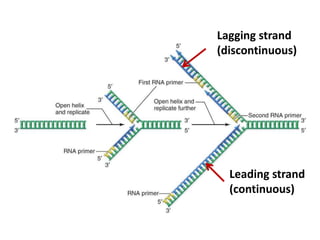
2) DNA replicates semi-conservatively, with each parent strand serving as a template to produce two new DNA double helices.
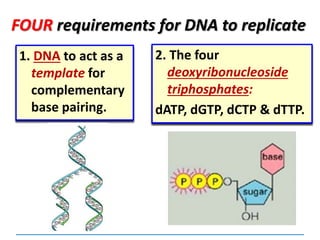
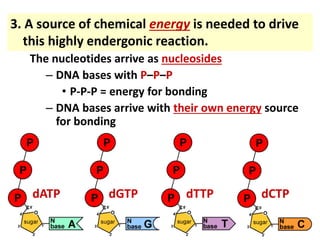
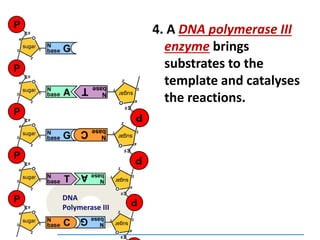
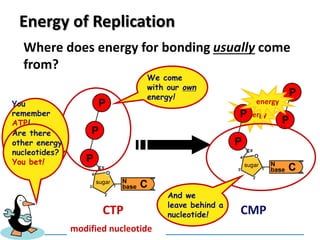
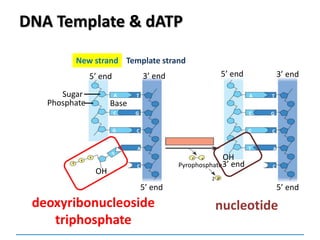
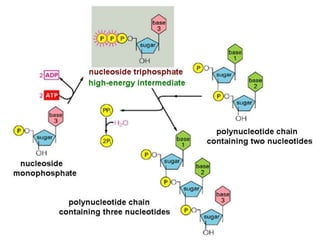
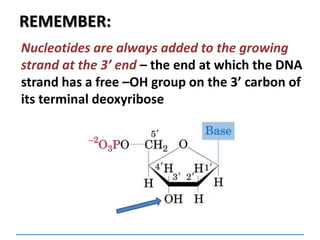
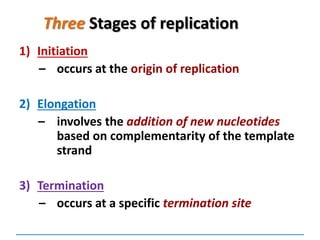
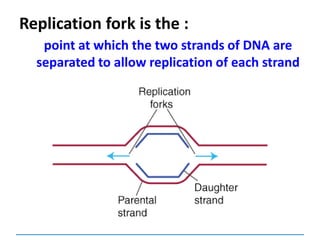
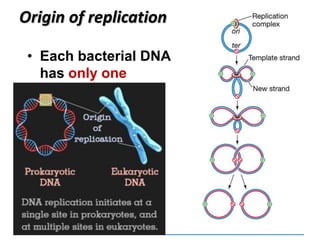
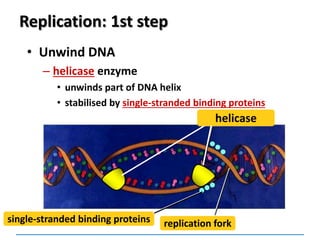
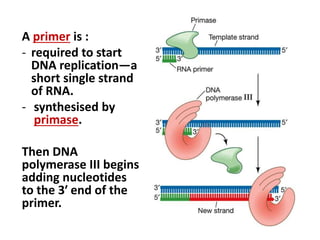
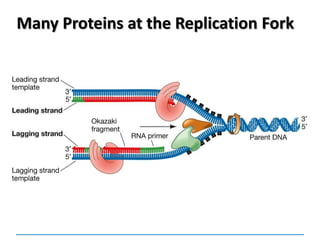
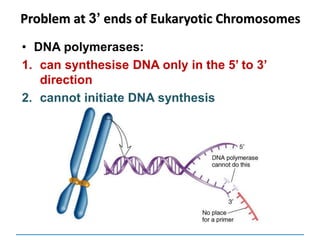
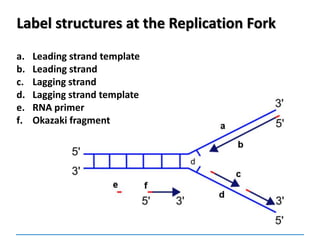
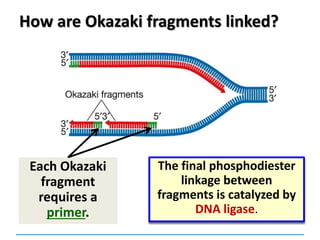
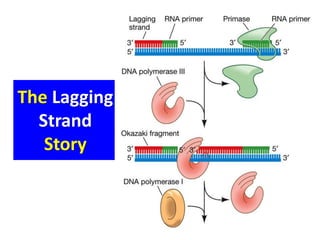
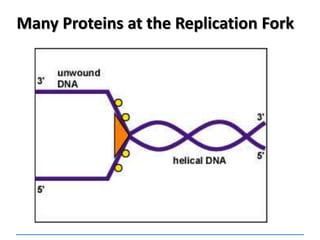
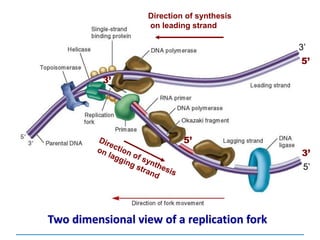
3) Replication requires DNA polymerase, nucleotides, and energy and proceeds through initiation, elongation, and termination steps at the replication fork.


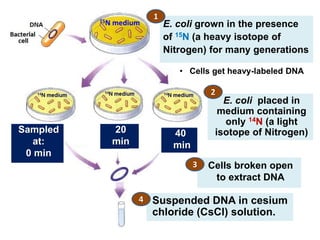
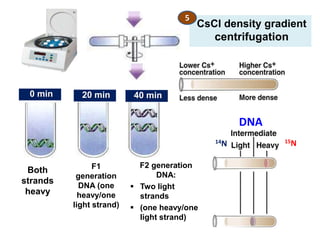
![Meselson and Stahl experiment
[1958] demonstrates
semiconservative replication:](https://image.slidesharecdn.com/dnareplication-141218092042-conversion-gate02-150523182342-lva1-app6891/85/Dnareplication-16-320.jpg)
![Protein Role
DNA helicases Unwinds the double helix
RNA primase Synthesises RNA primers
Single-strand binding
proteins
Keep the two strands separated
DNA polymerase I Erases primer and fills gaps
DNA polymerase II
[not in syllabus]
Proofreading of DNA
DNA polymerase III Synthesises DNA; proofreading
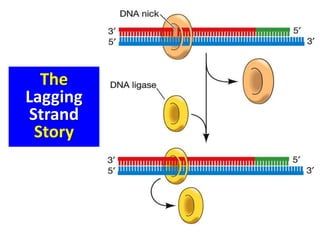
DNA ligase Joins the ends of DNA segments;
DNA repair](https://image.slidesharecdn.com/dnareplication-141218092042-conversion-gate02-150523182342-lva1-app6891/85/Dnareplication-41-320.jpg)



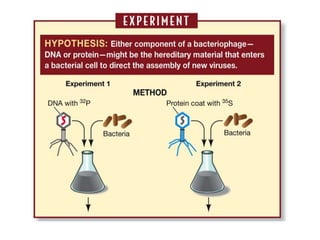
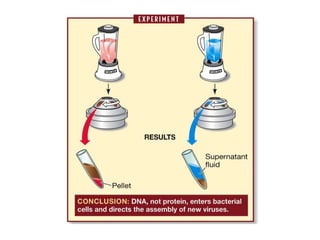
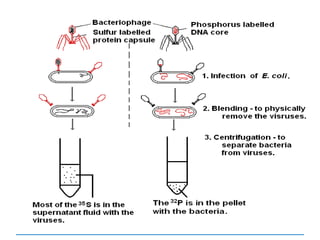
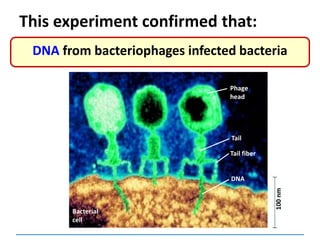

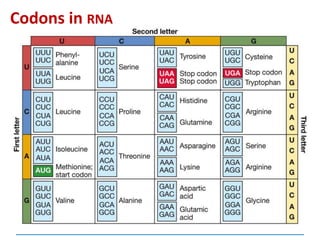
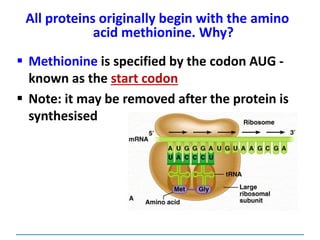
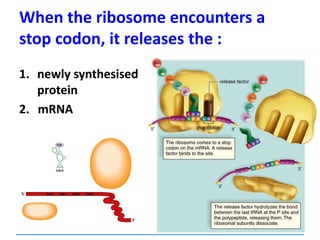
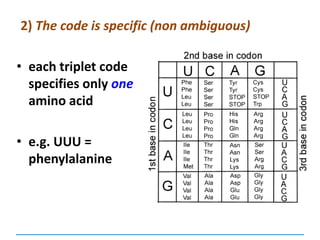
![3) The code is degenerate
Valine
GUU
GUC
GUA
GUG
a given amino acid may be coded for by more
than one codon
64 codons and only 20
amino acids:
so some amino acids
are coded for by
several codons –
exceptions [next
slide]:
Tyrosine
UAU
UAC
Lysine
AAA
AAG](https://image.slidesharecdn.com/dnareplication-141218092042-conversion-gate02-150523182342-lva1-app6891/85/Dnareplication-97-320.jpg)
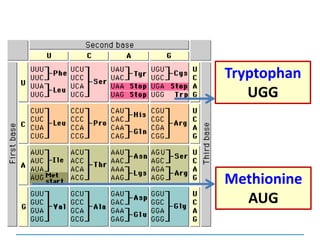
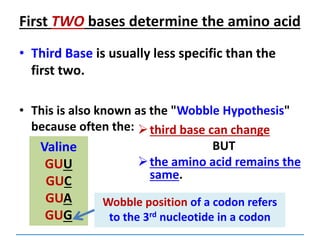
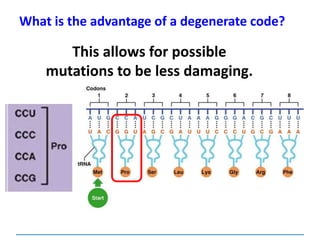




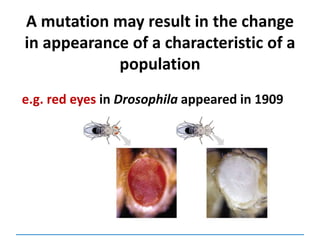

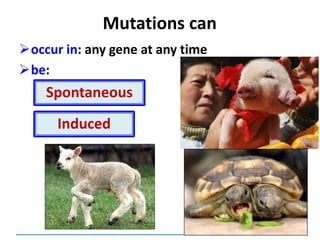
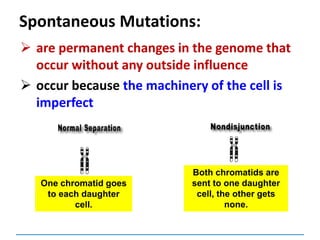
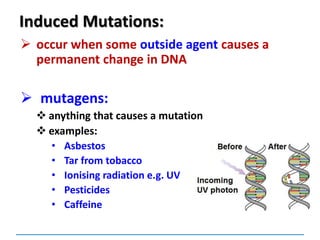

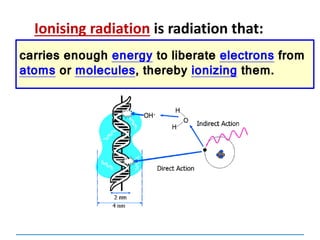
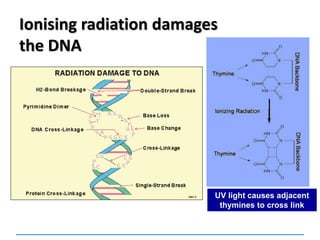
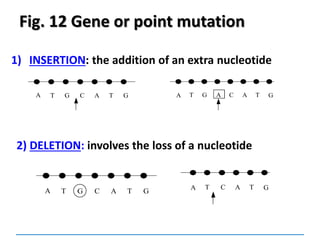
![Mutations
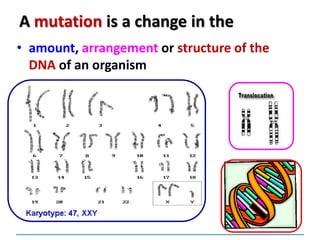
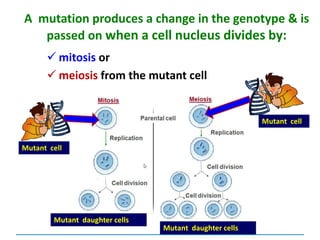
can be:
Chromosomal
[covered in 2nd year]
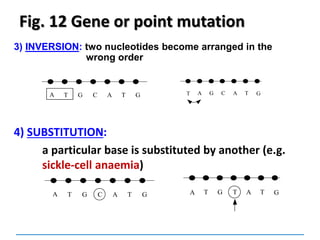
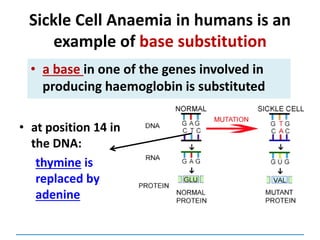
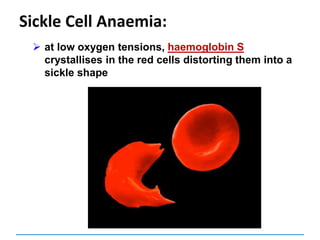
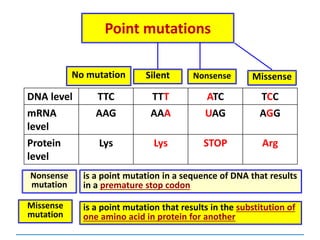
Gene mutations or point mutations:
INSERTION
INVERSION
DELETION
SUBSTITUTION
describe a change in the structure of DNA
at a single locus
1
2](https://image.slidesharecdn.com/dnareplication-141218092042-conversion-gate02-150523182342-lva1-app6891/85/Dnareplication-130-320.jpg)

![End-Of-Year SEP 2013
Use your knowledge of the genetic code to explain
statements (a) and (b) below. Use your knowledge
of genetic mutations to answer statements (c), (d)
and (e). [5 marks each]
i) Distinguish between a base substitution and an
inversion.
i) Distinguish between a deletion and an insertion.
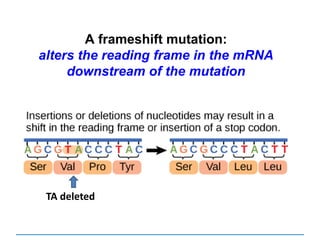
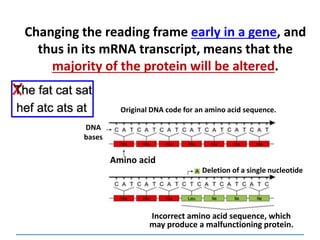
ii) Explain how deletions and insertions lead to
frameshift mutations](https://image.slidesharecdn.com/dnareplication-141218092042-conversion-gate02-150523182342-lva1-app6891/85/Dnareplication-139-320.jpg)
![Use your knowledge of biology to explain the following.
The structure of the DNA molecule permits vast amounts
of information to be stored. (5 marks)
Question: [SEP, 2007]
1. Information on the DNA molecule is in the form of a
sequence of bases, where three consecutive bases
specify an amino acid. Thus a small number of bases are
needed to code for an amino acid. Considering that DNA
within a eukaryotic cell is 2m long, it allows for a large
amount of information to be stored.
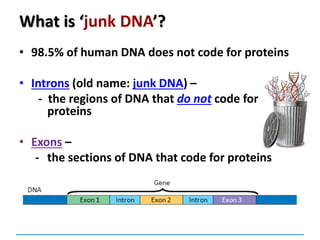
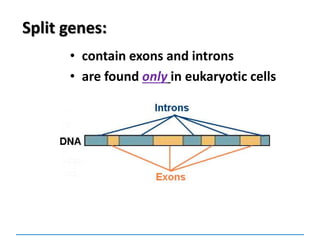
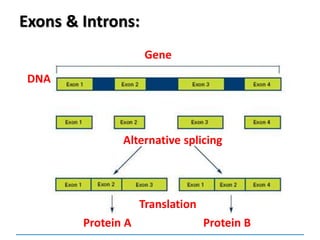
2. In many eukaryotic cells, split genes occur. These
contain regions which code for the protein called exons
and introns which do not code. The way in which the
exons are linked together determines the type of
polypeptide to be formed. Thus one gene can form a
number of closely related polypeptides.](https://image.slidesharecdn.com/dnareplication-141218092042-conversion-gate02-150523182342-lva1-app6891/85/Dnareplication-140-320.jpg)