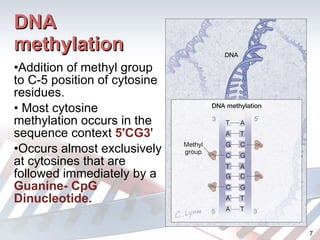
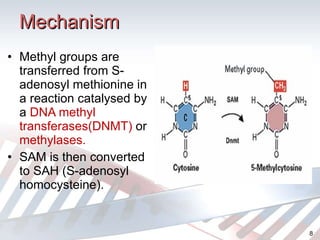
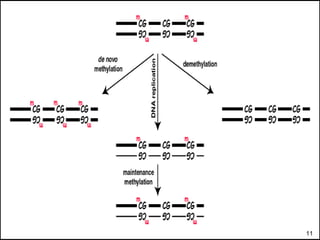
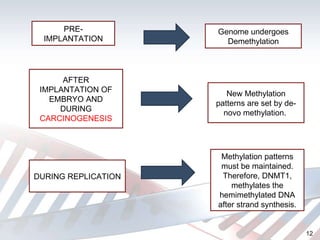
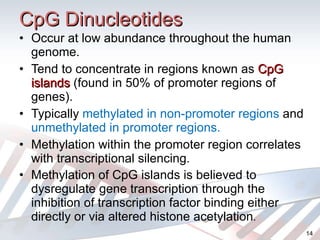
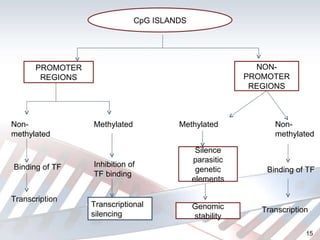
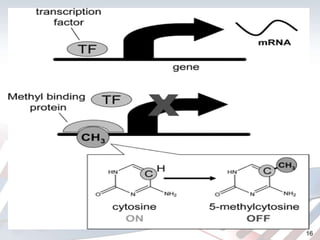
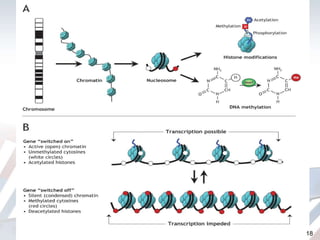
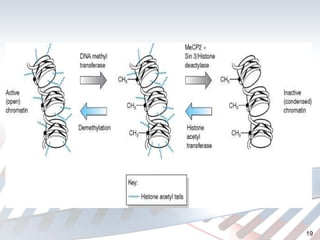
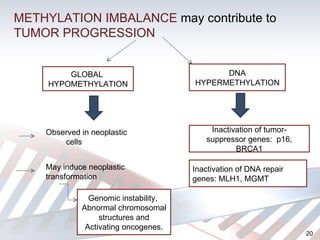
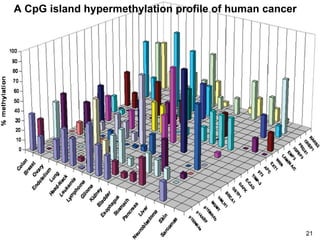
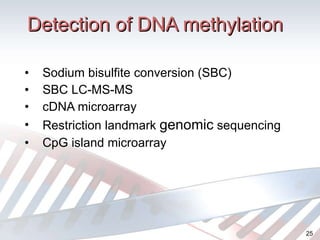
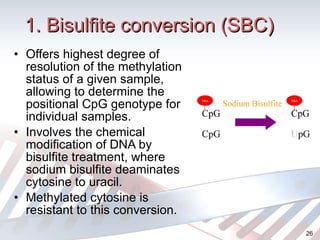
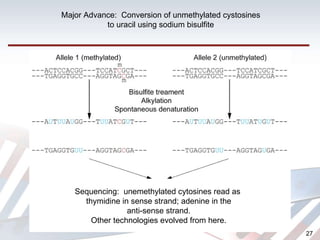
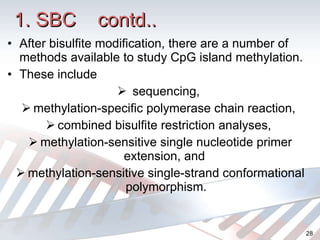
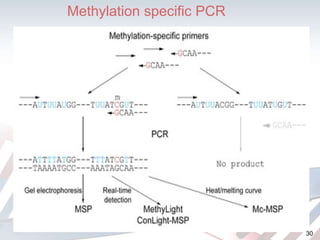
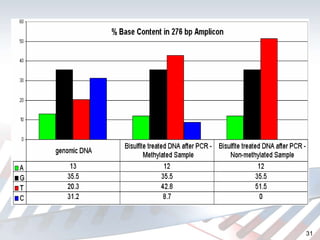
DNA methylation is an epigenetic mechanism that involves the addition of a methyl group to cytosine residues in DNA. It is catalyzed by DNA methyltransferase enzymes and plays a key role in gene expression and cellular differentiation. Aberrant DNA methylation, including both hypermethylation and hypomethylation, has been associated with cancer development by disrupting gene expression. Detection of DNA methylation patterns can provide insights into cancer biology and may have applications as a diagnostic tool.