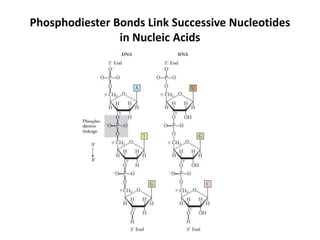
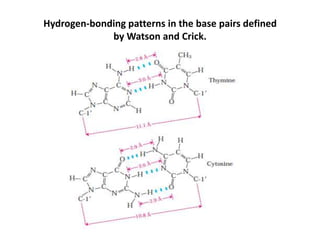
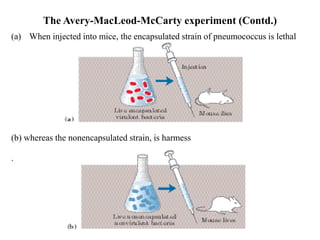
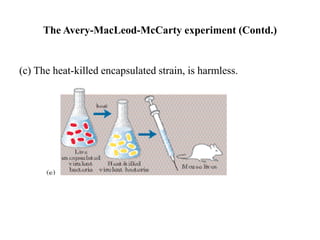
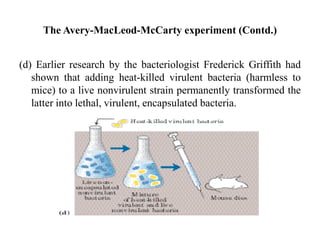
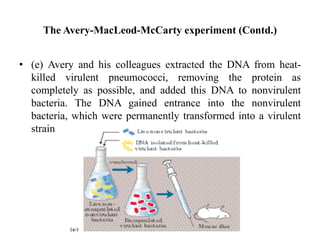
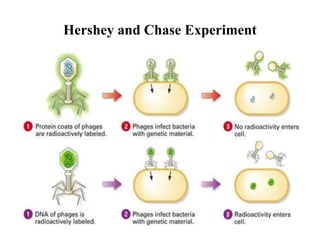
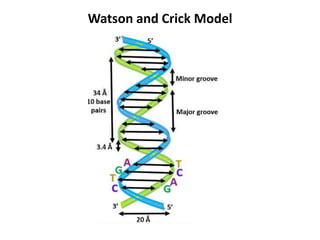
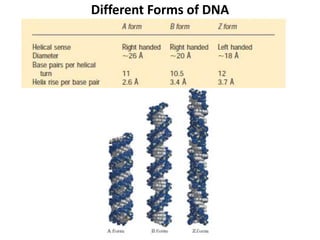
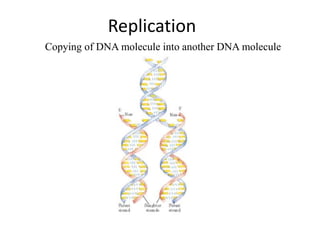
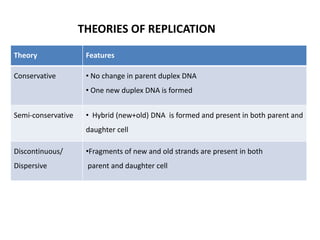
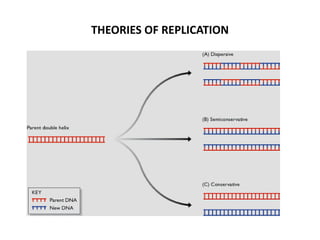
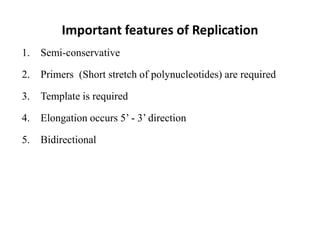
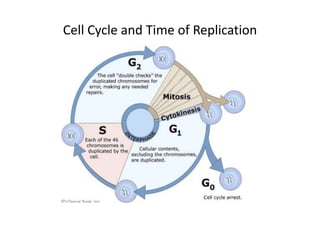
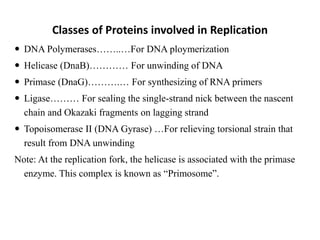
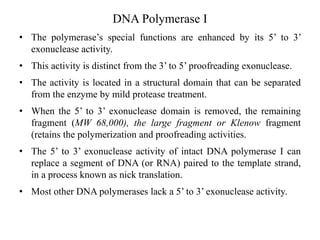
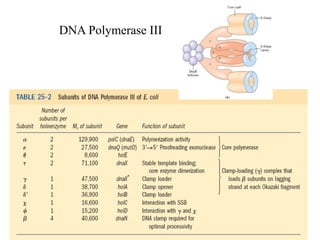
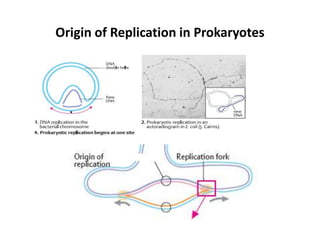
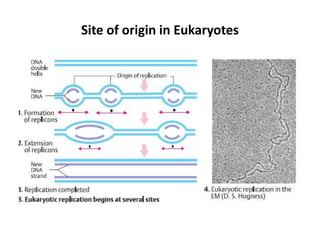
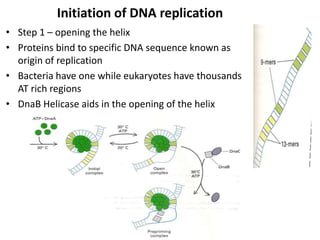
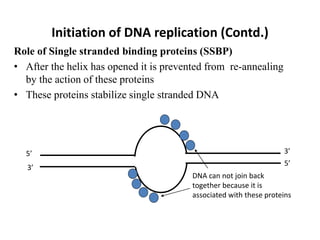
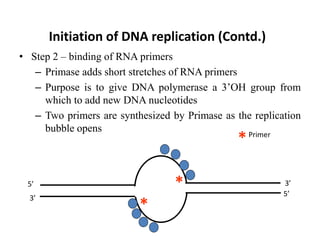
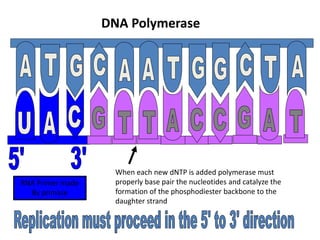
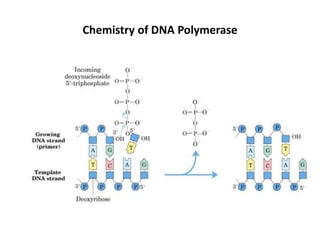
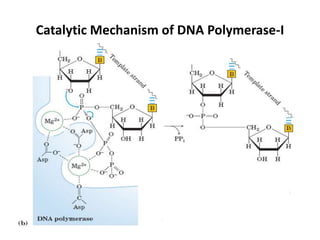
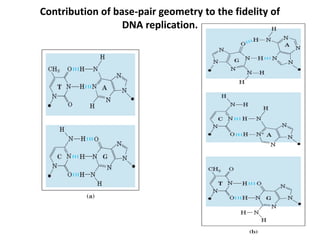
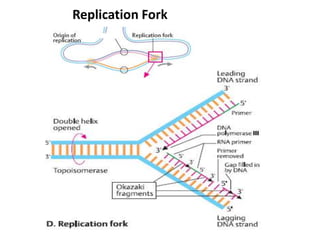
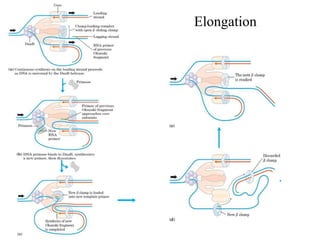
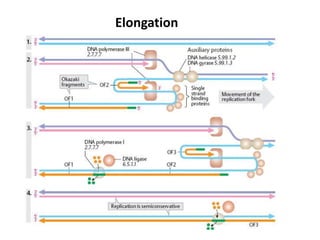
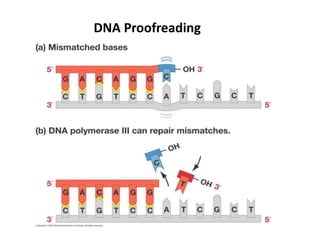
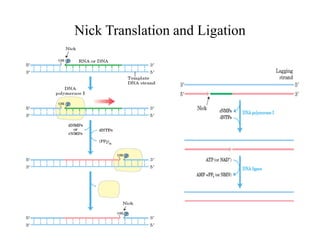
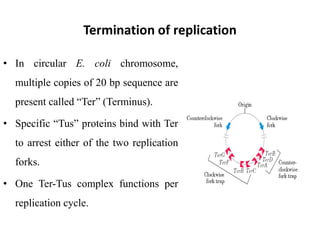
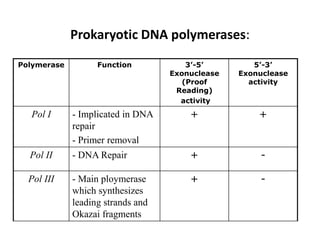
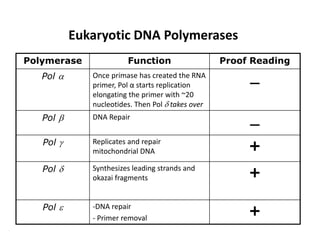
The document discusses DNA replication. It describes early experiments that showed DNA carries genetic information, such as the Avery-MacLeod-McCarty experiment. It also describes Chargaff's rules about DNA base composition and the Watson and Crick model of the DNA double helix structure. The process of DNA replication is then explained, including semi-conservative replication, the role of enzymes like DNA polymerase and helicase, and leading and lagging strand synthesis.