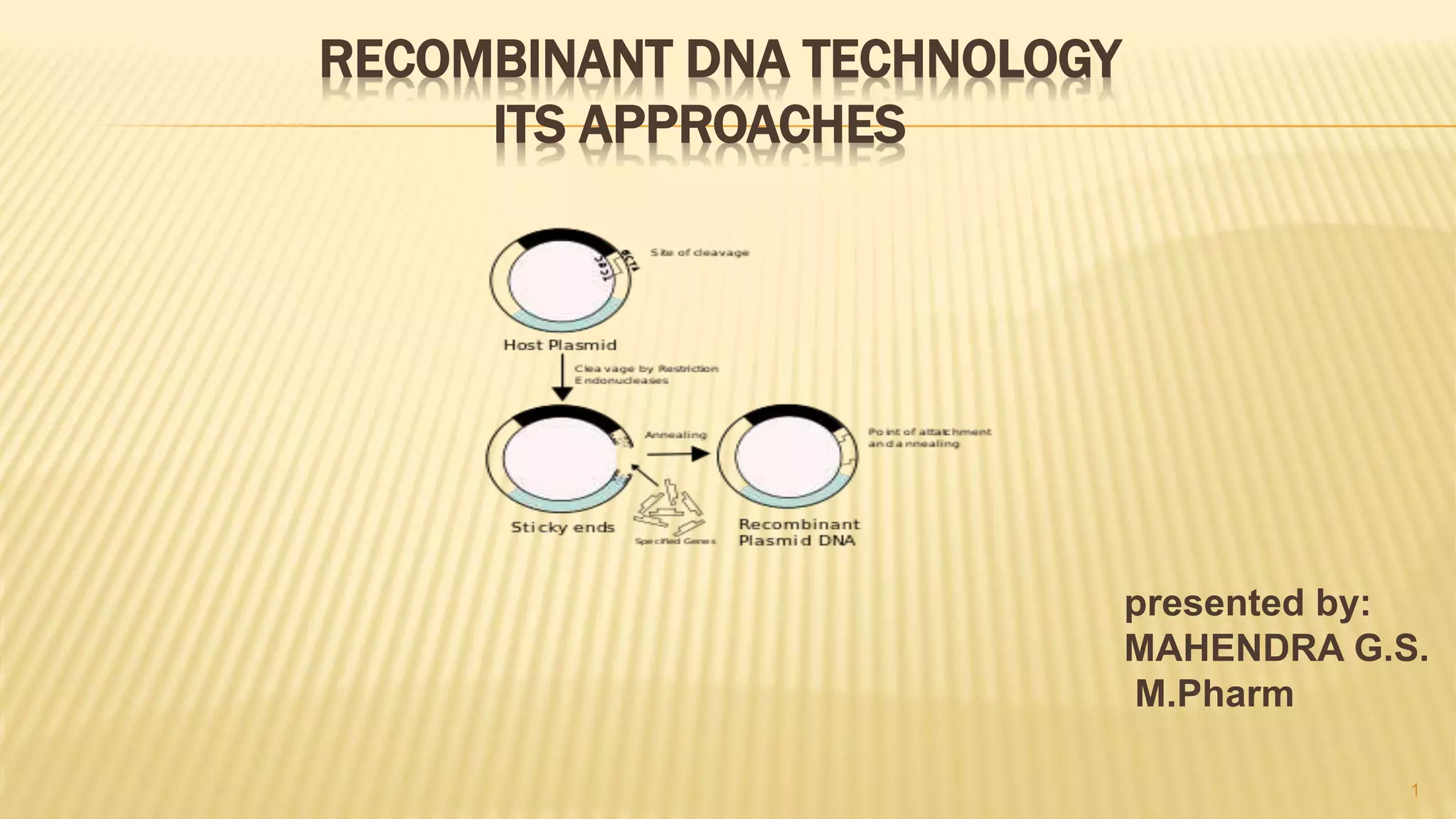
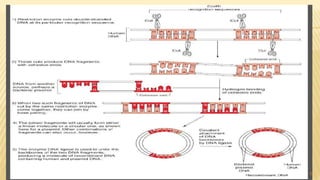
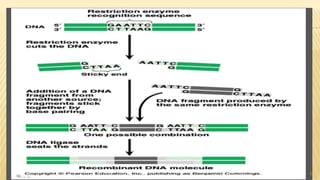
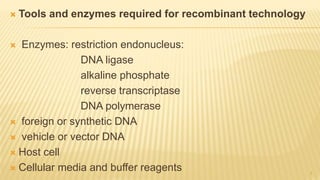
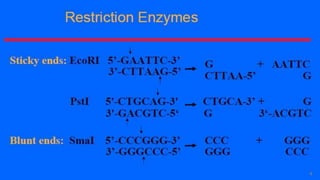
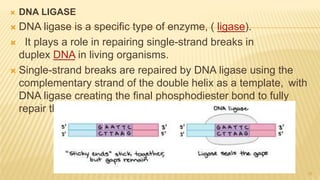
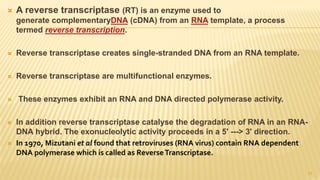
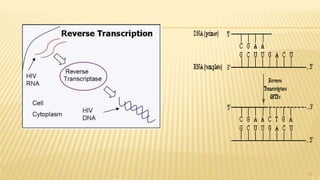
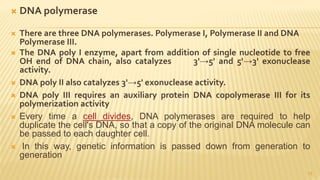
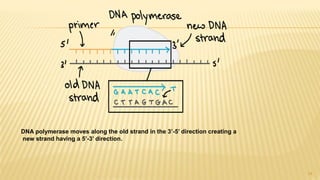
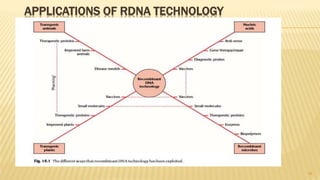
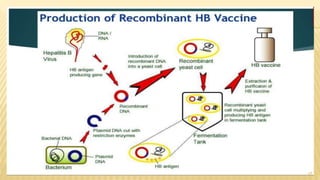
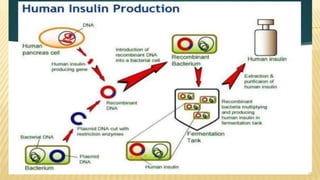
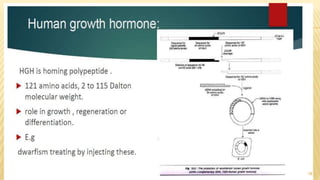
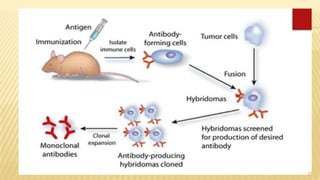
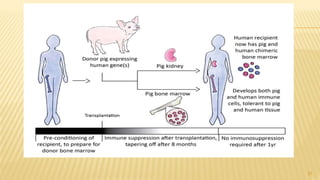
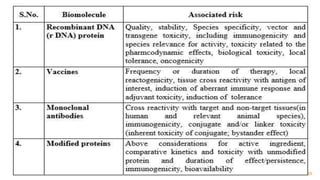
This document discusses recombinant DNA technology and its applications. It begins with an introduction to recombinant DNA technology and its history. It then describes the tools and enzymes used, including restriction enzymes, DNA ligase, reverse transcriptase, and DNA polymerase. Various vectors like bacterial plasmids and bacteriophages are also discussed. The document outlines several applications such as monoclonal antibody production, disease diagnosis, DNA fingerprinting, environmental uses, gene therapy, and xenotransplantation. In summary, the document provides an overview of the key concepts, techniques, and uses of recombinant DNA technology.