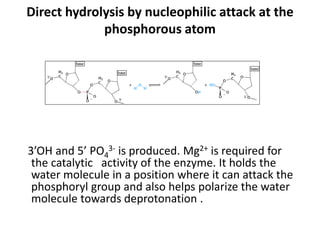
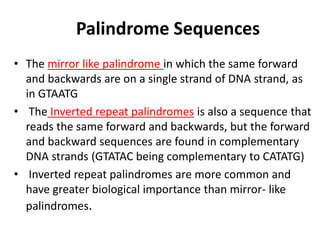
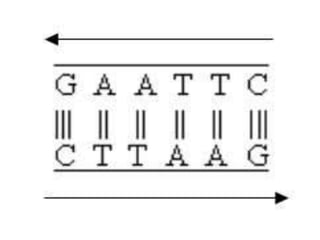
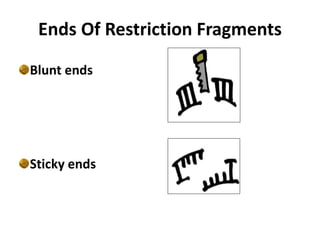
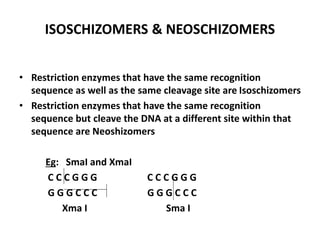
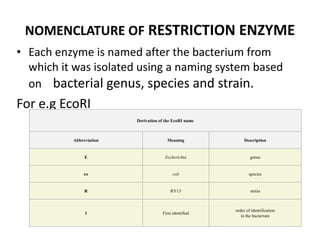
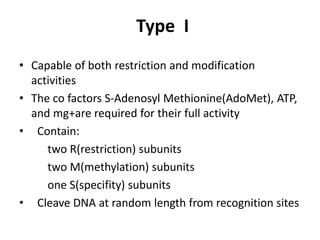
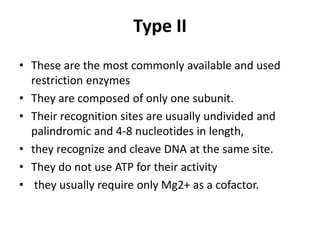
Restriction enzymes are molecular scissors found in bacteria that cut DNA molecules at specific recognition sequences. They serve as a defensive mechanism for bacteria against bacteriophages by cleaving the phage DNA. There are over 3000 known restriction enzymes that are classified into four main types based on their composition, cofactors, and cutting mechanisms. Restriction enzymes are important tools in biotechnology for manipulating DNA sequences through cutting DNA fragments with specific sticky or blunt ends, which can then be recombined through techniques like cloning.