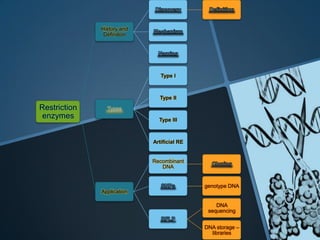
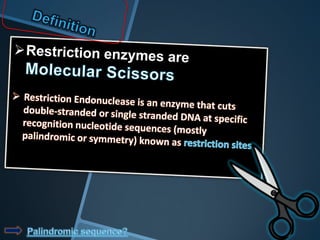
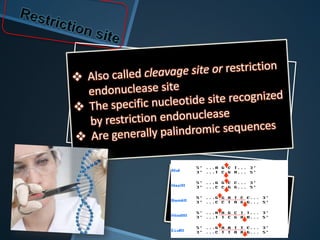
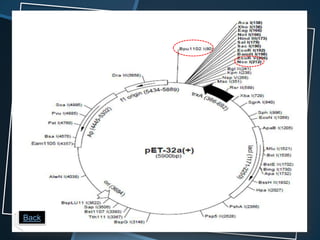
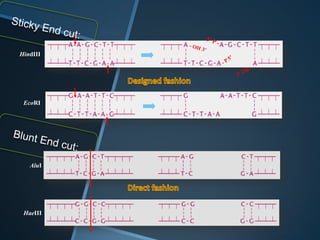
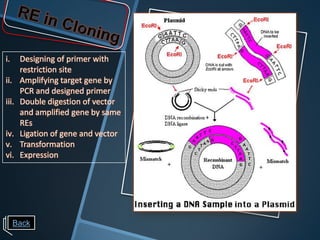
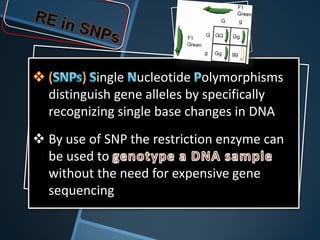
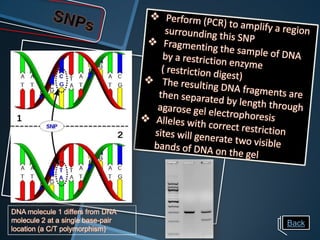
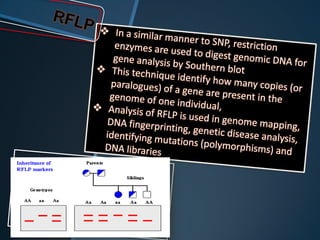
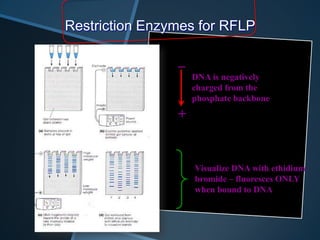
Restriction enzymes are enzymes that cut DNA at specific recognition sequences. There are three main types of restriction enzymes - Type I, II, and III. Type II restriction enzymes are the most commonly used in molecular cloning and DNA manipulation. They recognize short palindromic DNA sequences of 4-8 base pairs and cleave DNA within or nearby these recognition sites. Restriction enzymes have many applications including DNA sequencing, DNA libraries, and recombinant DNA techniques which enable genetic engineering.