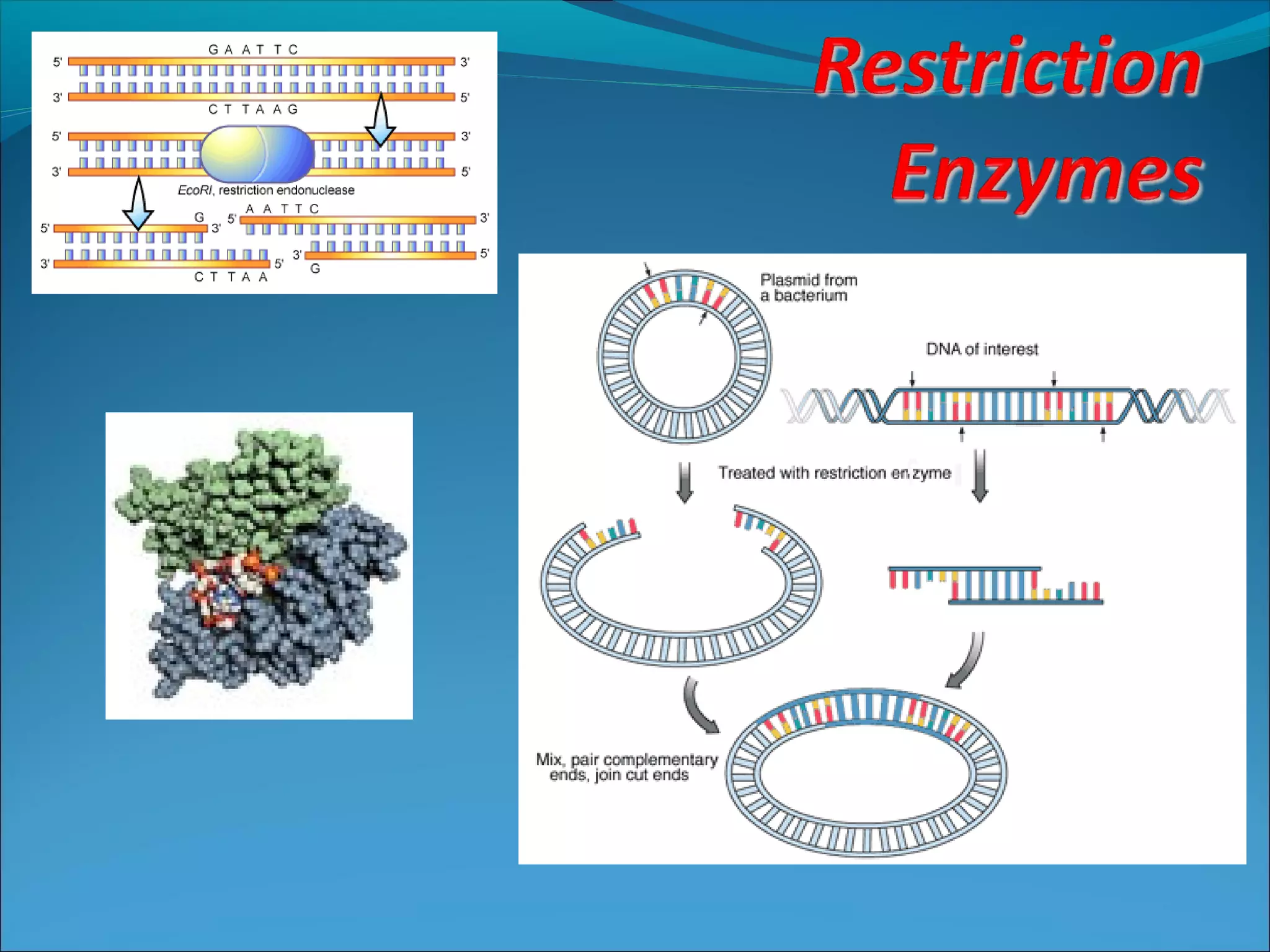
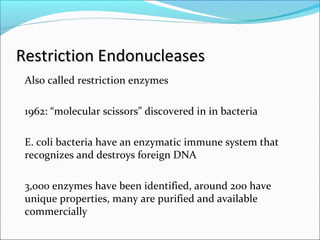
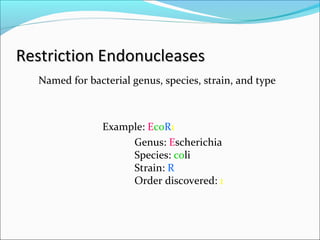
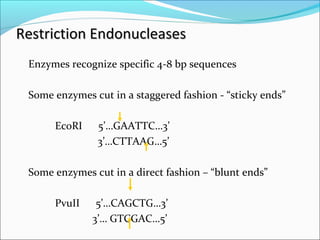
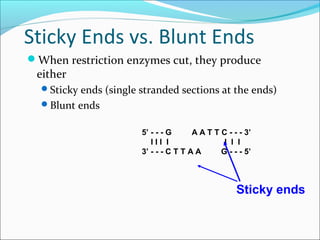
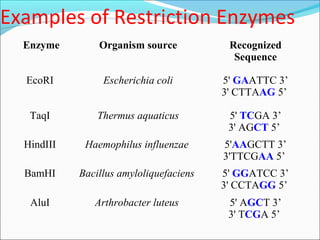
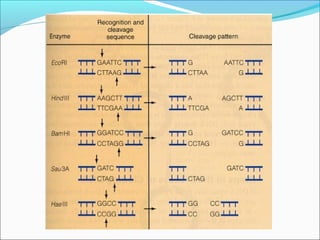
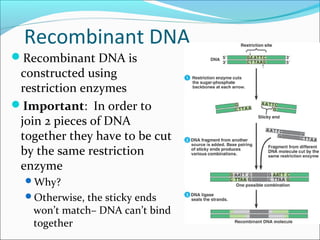
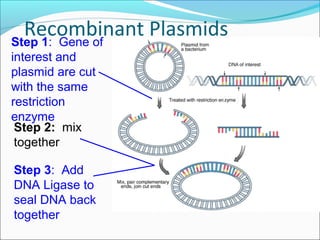
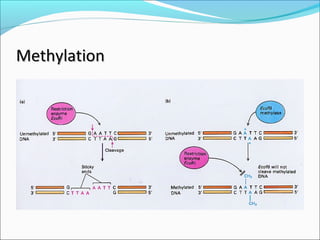
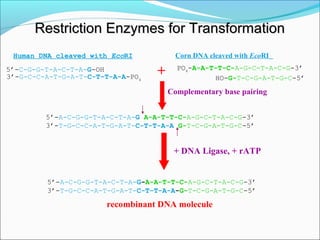
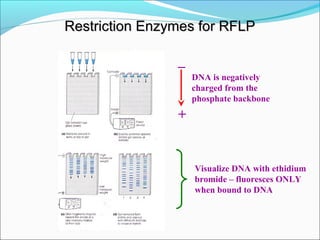
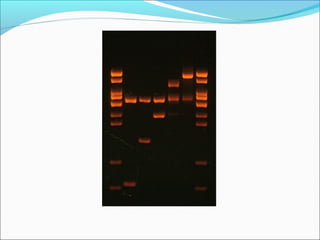
Restriction enzymes are molecular scissors found in bacteria that cut DNA at specific recognition sequences. They break the covalent bonds within a DNA strand and the hydrogen bonds between the strands. Bacteria use these enzymes to protect themselves from foreign DNA. Restriction enzymes are now used widely in laboratories to cut DNA in recombinant DNA techniques and other applications like DNA fingerprinting. They produce sticky or blunt ends that allow targeted joining of DNA fragments.