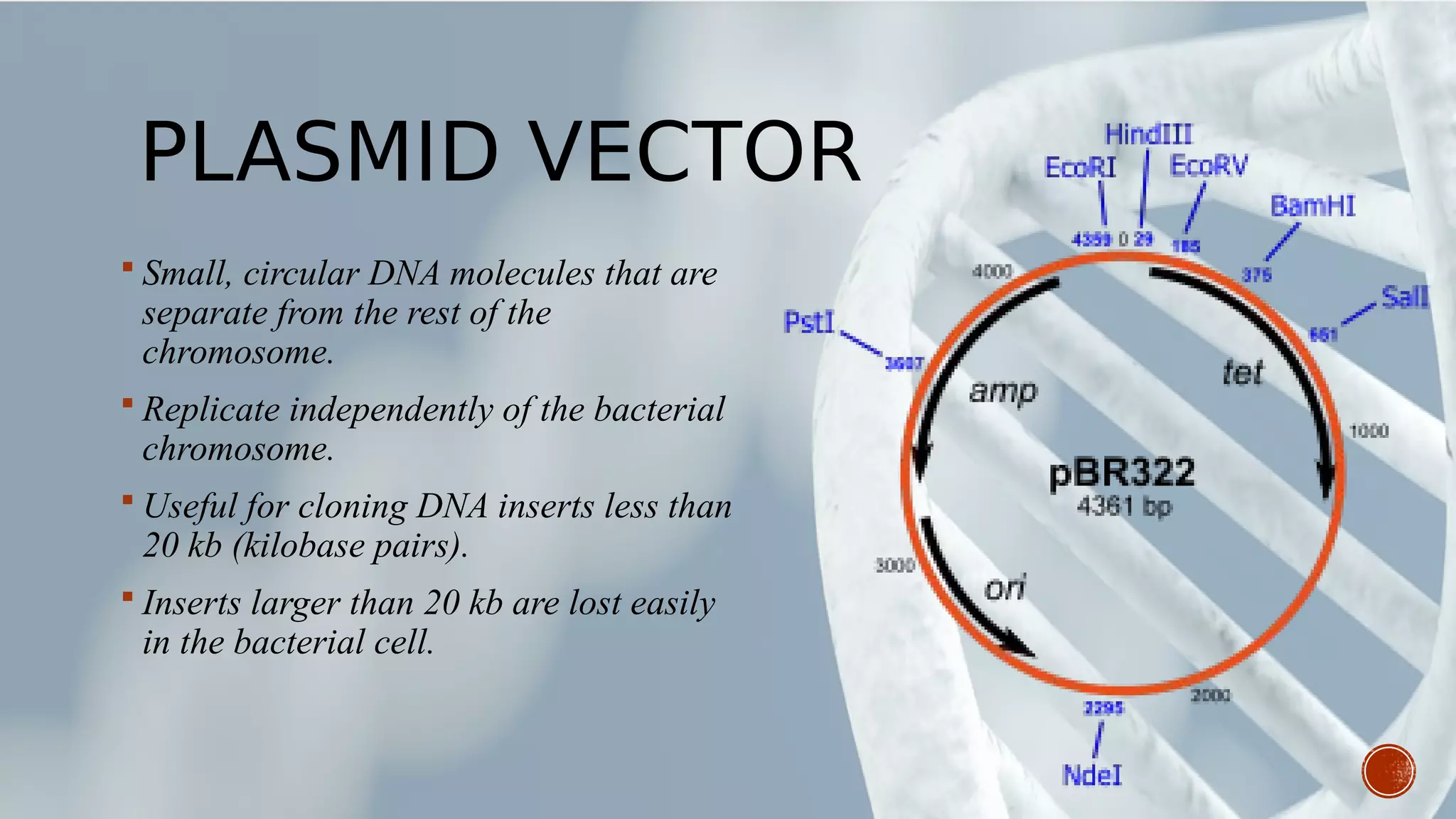
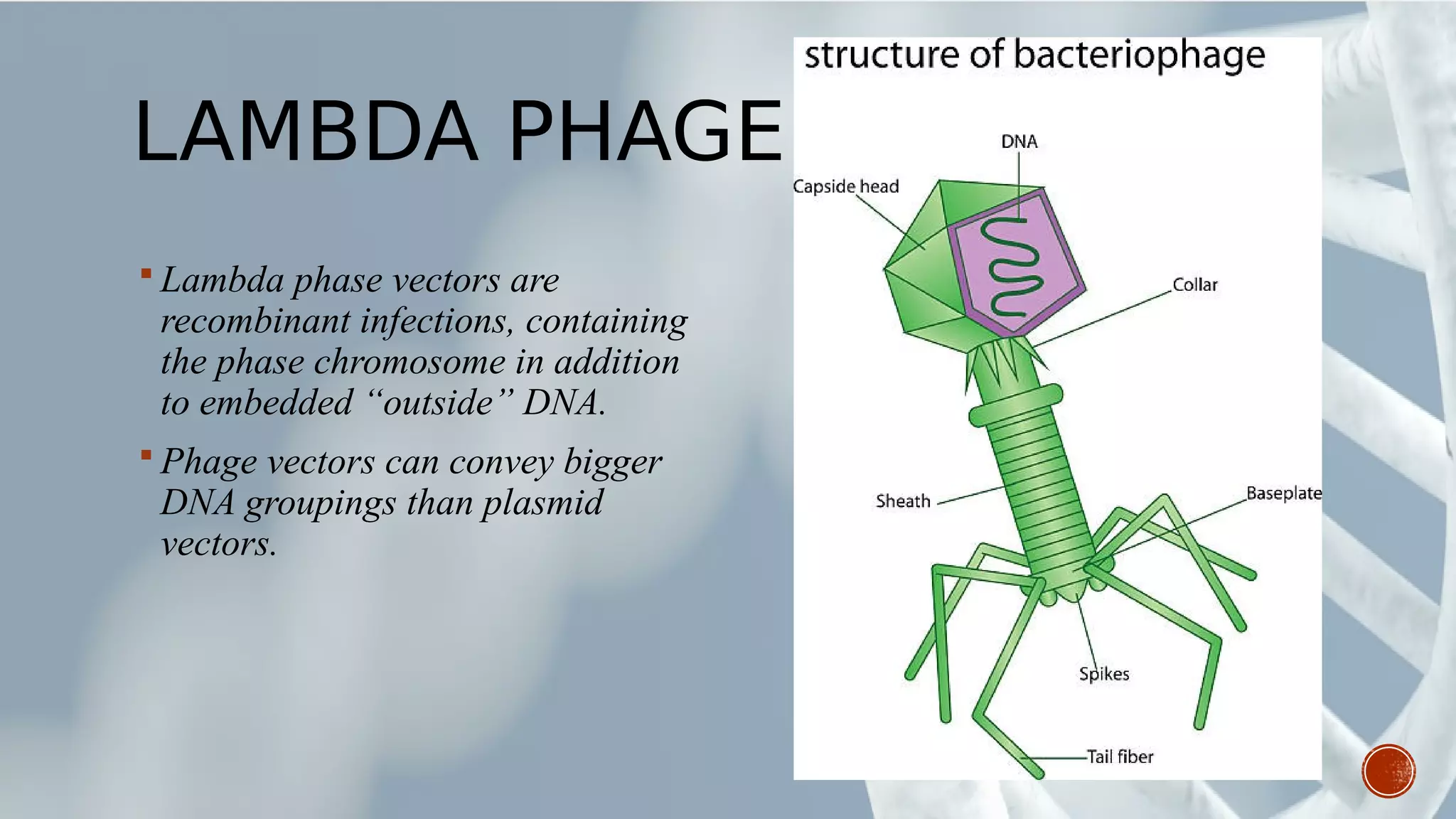
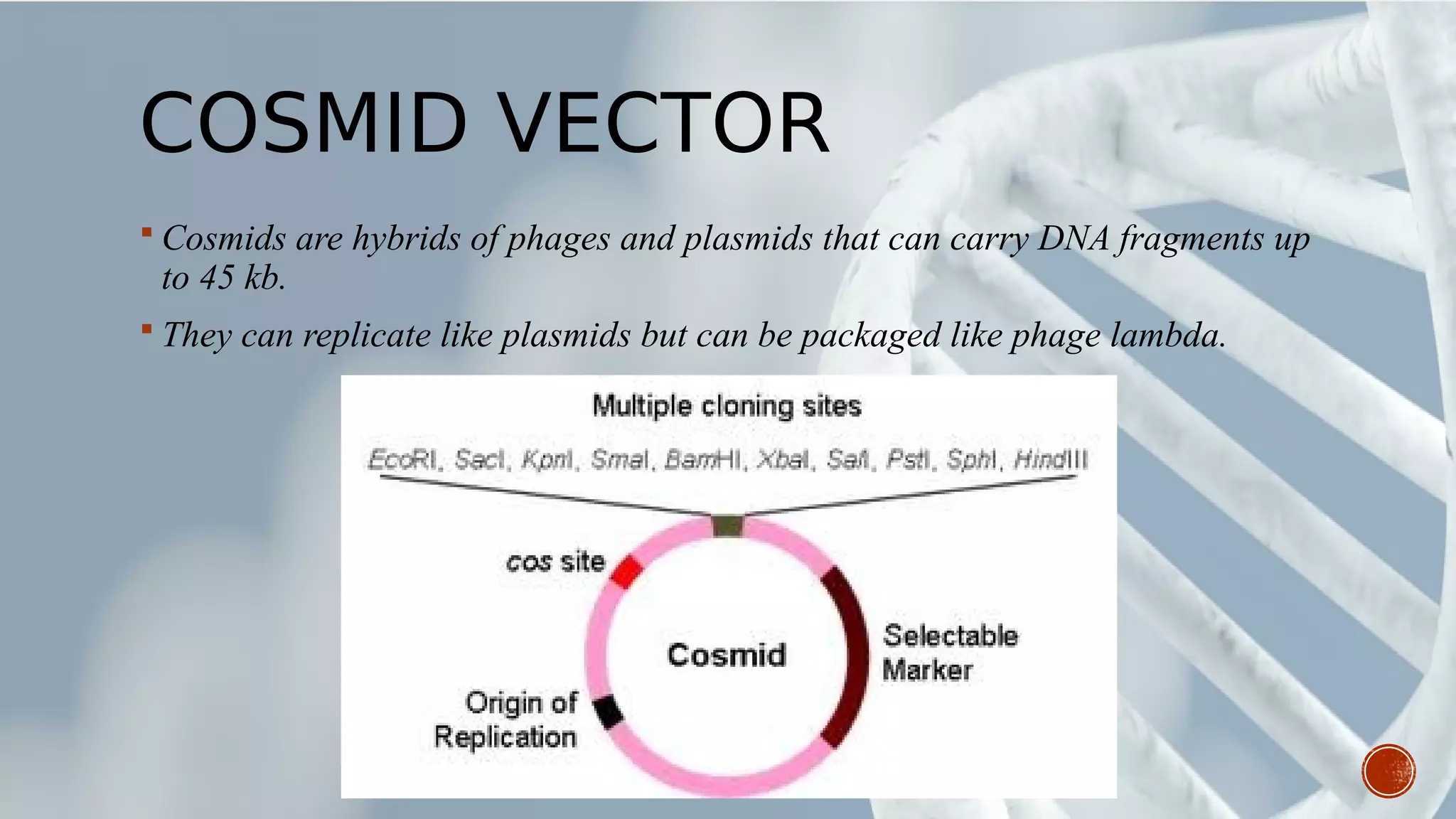
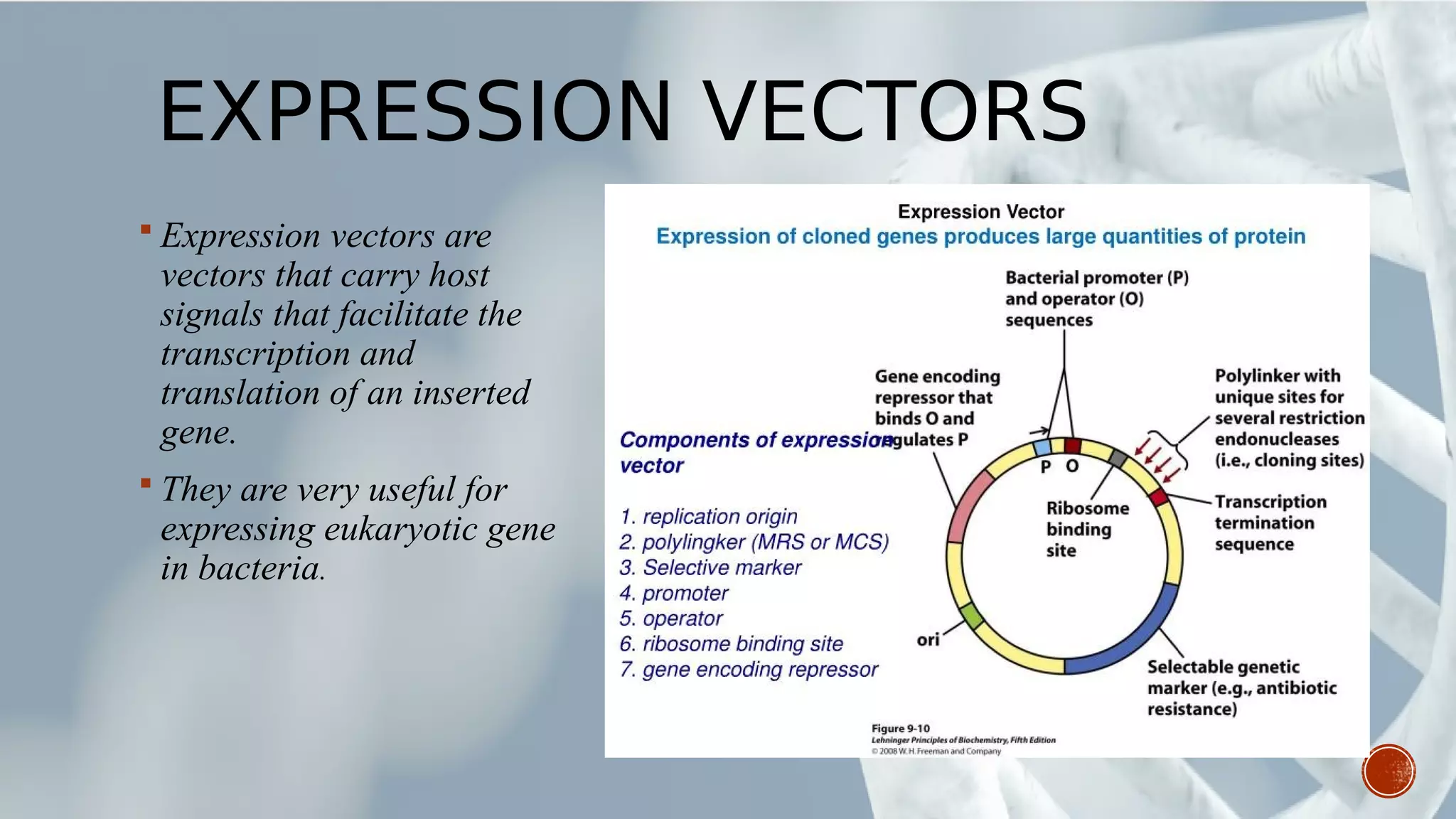
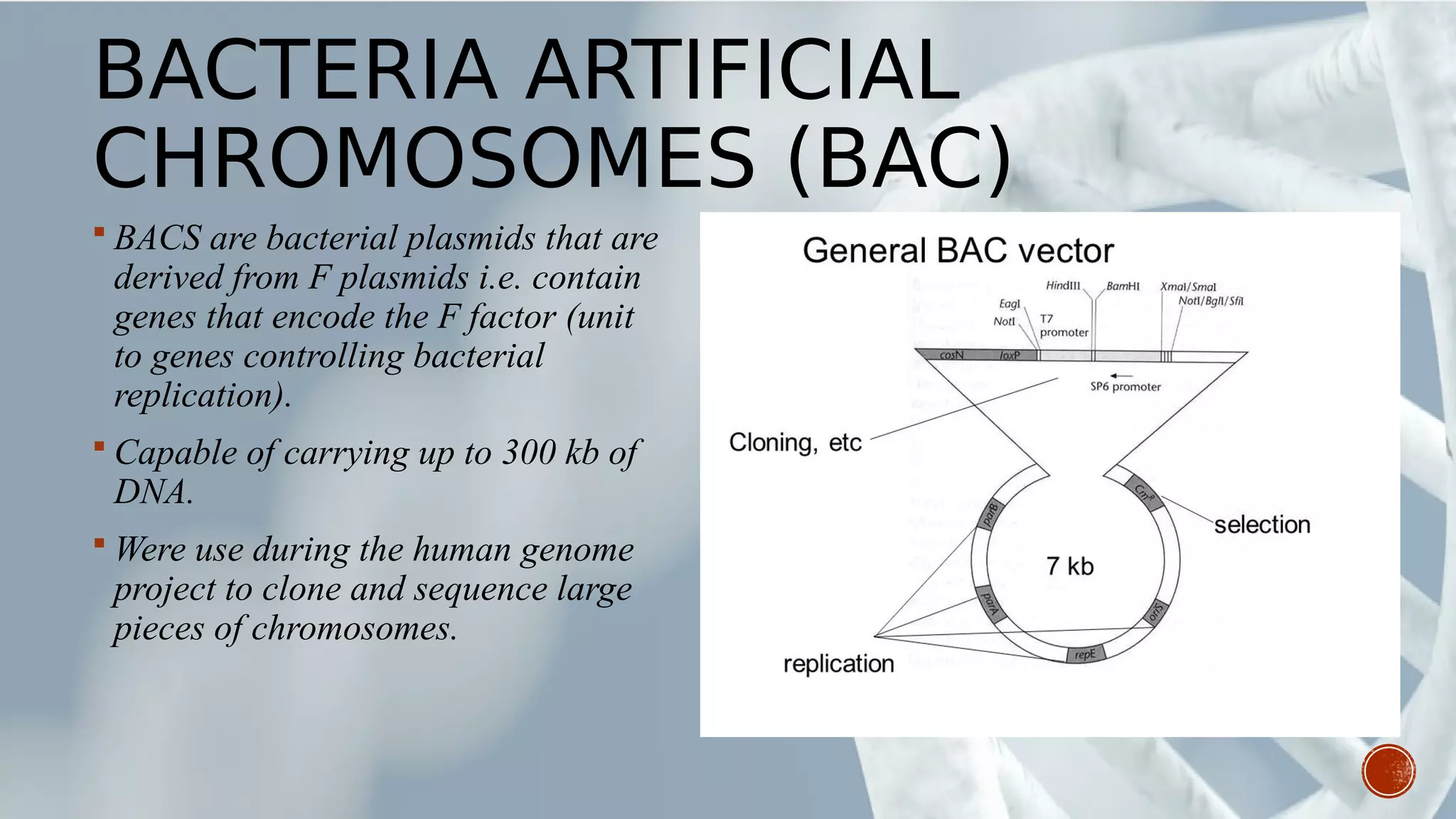
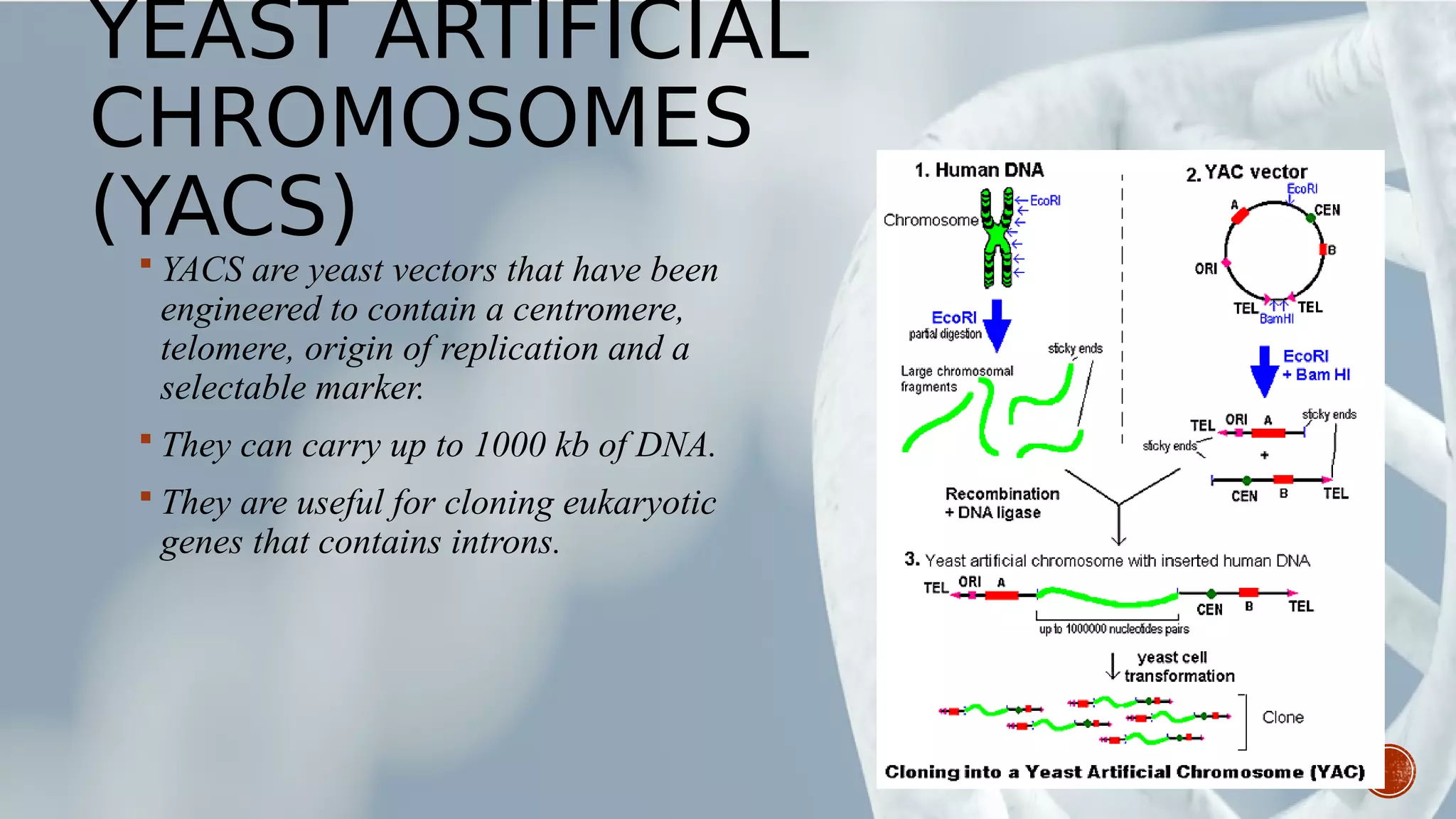
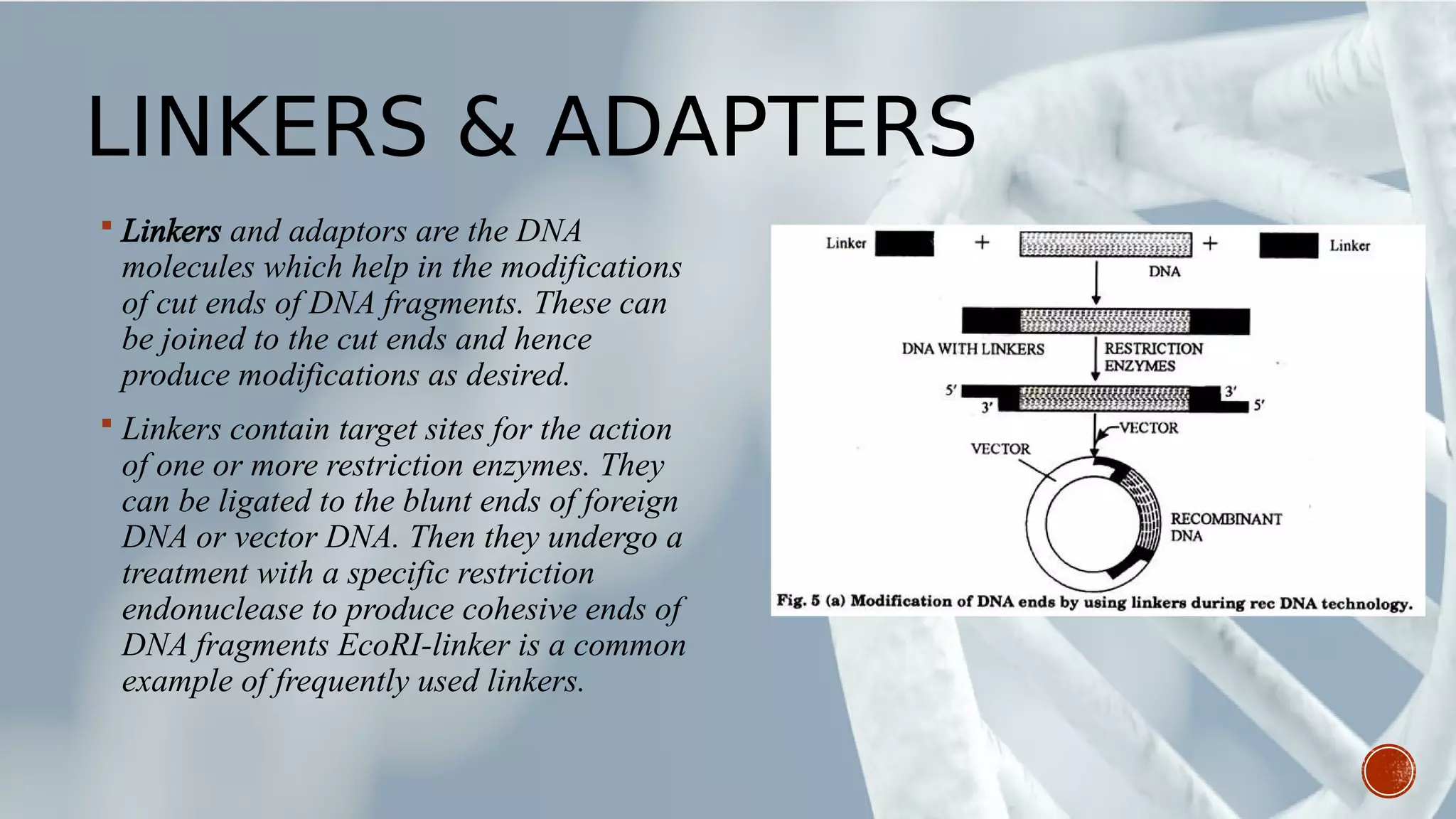
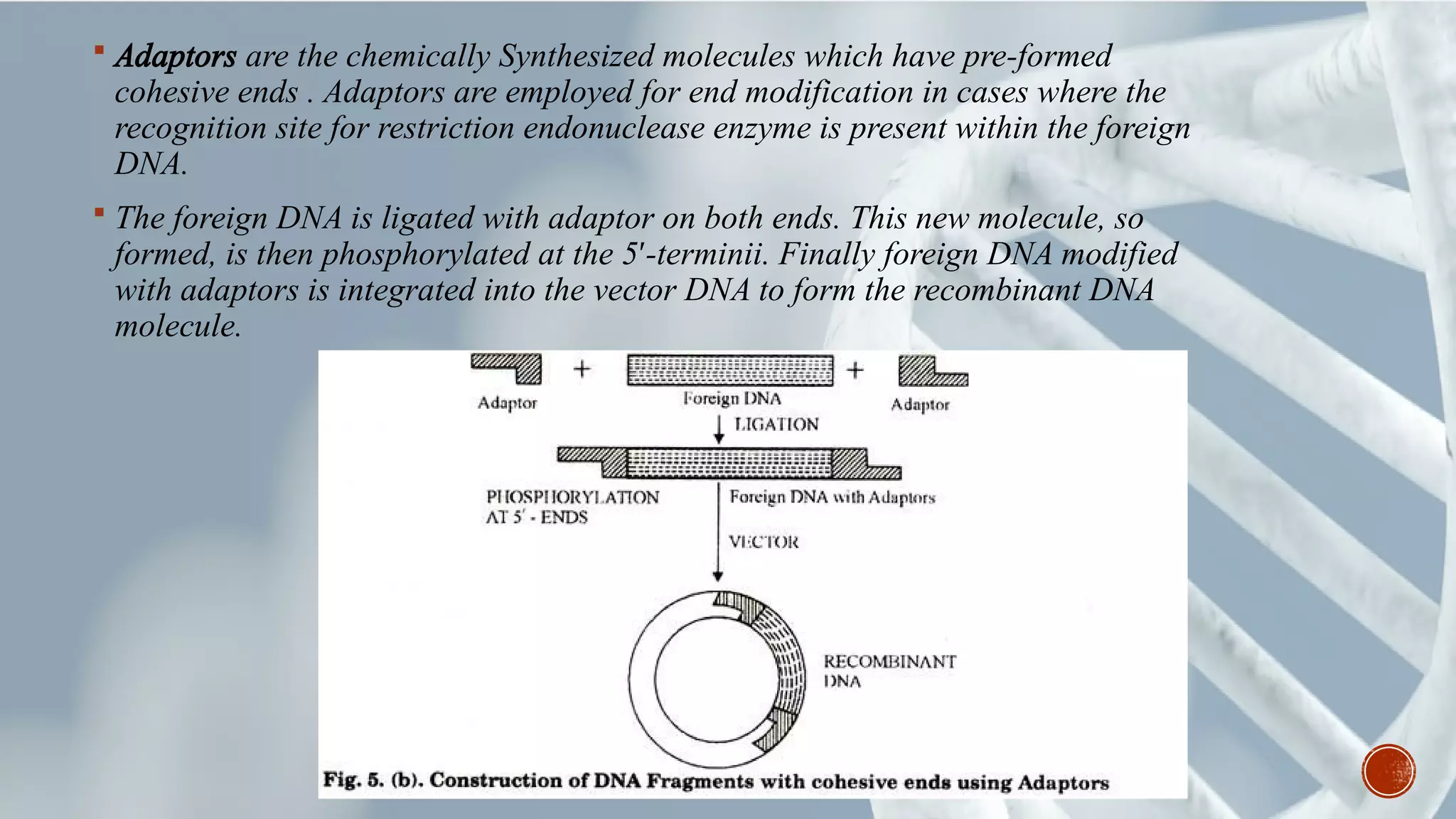
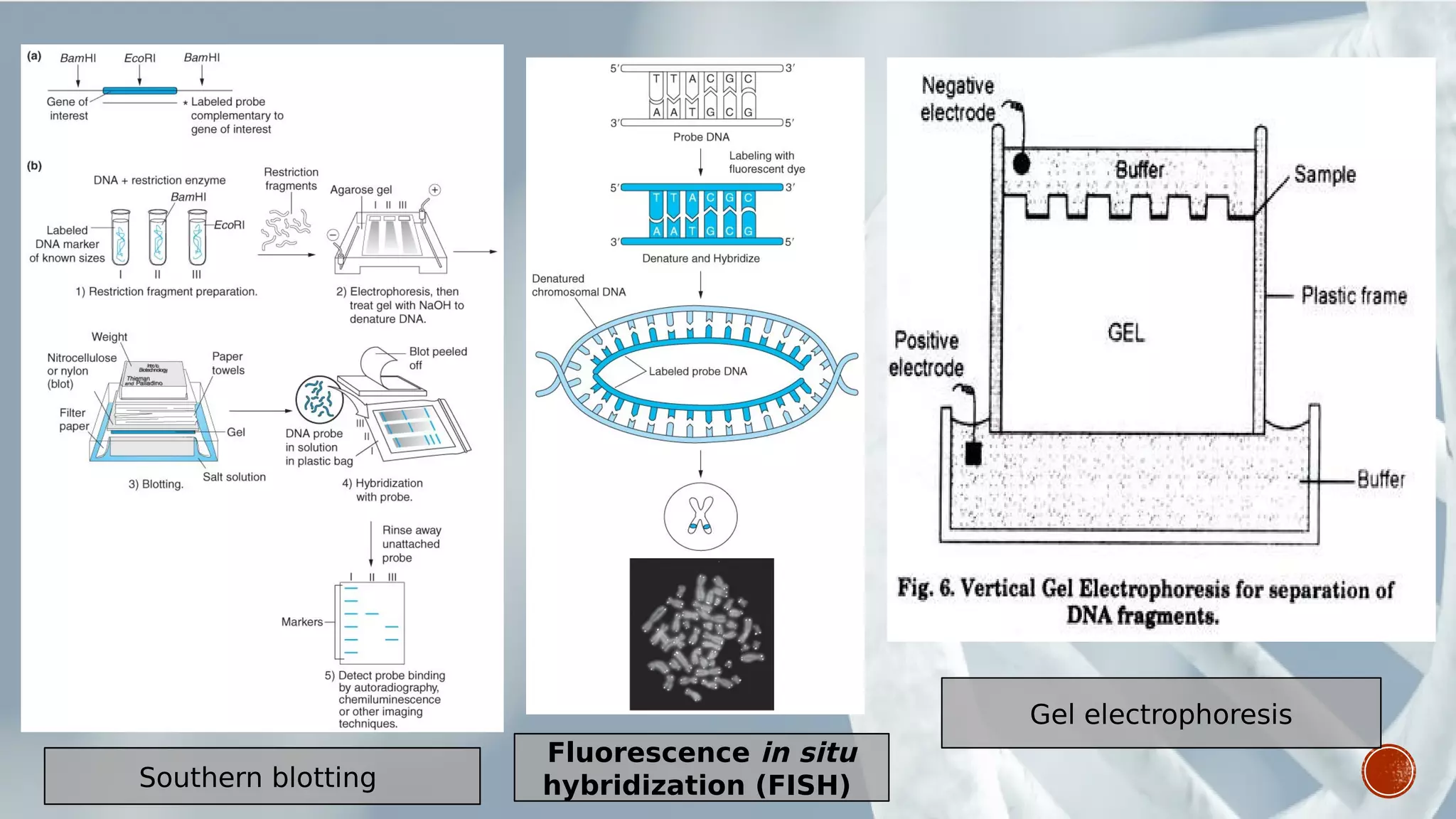
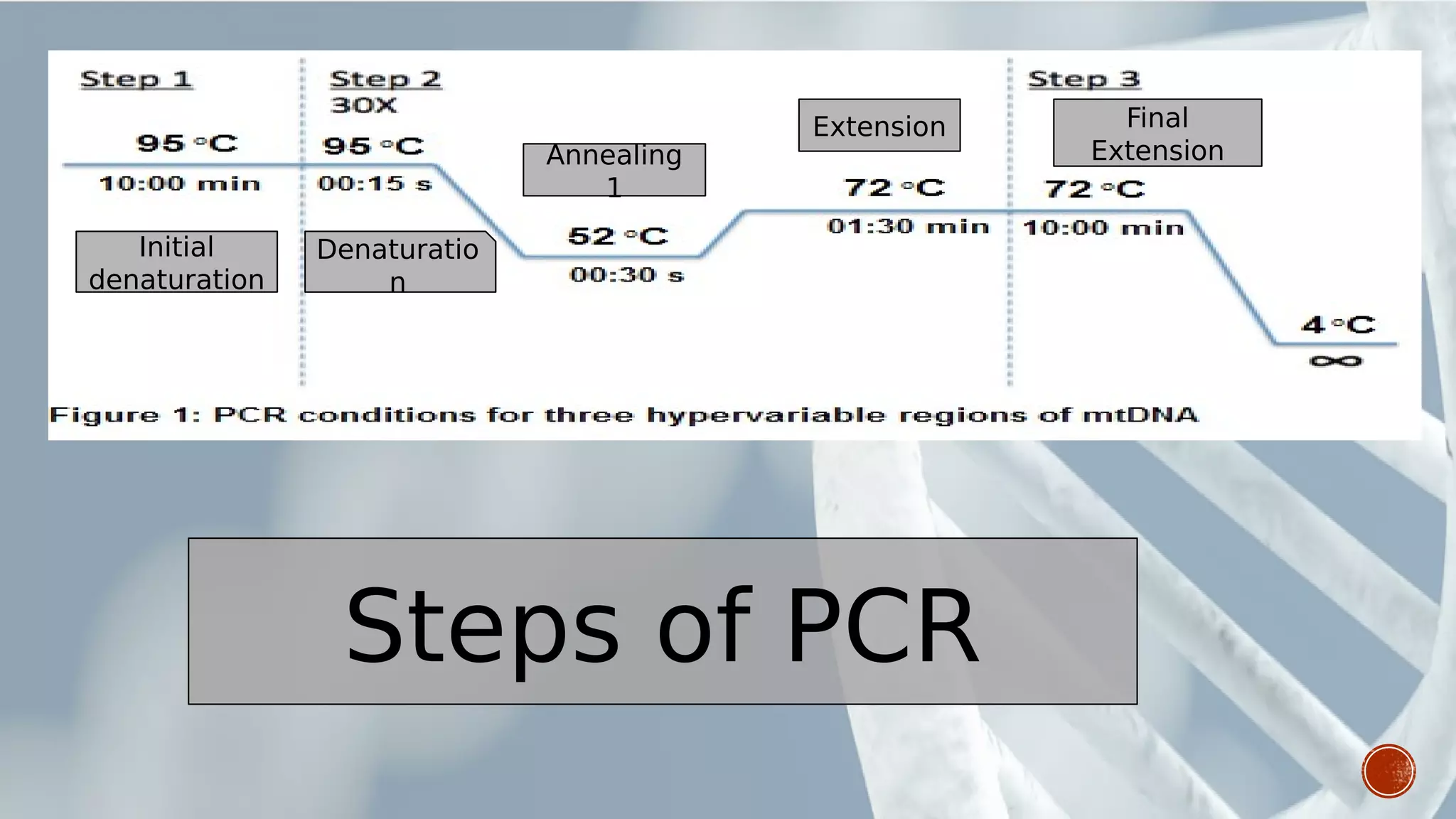
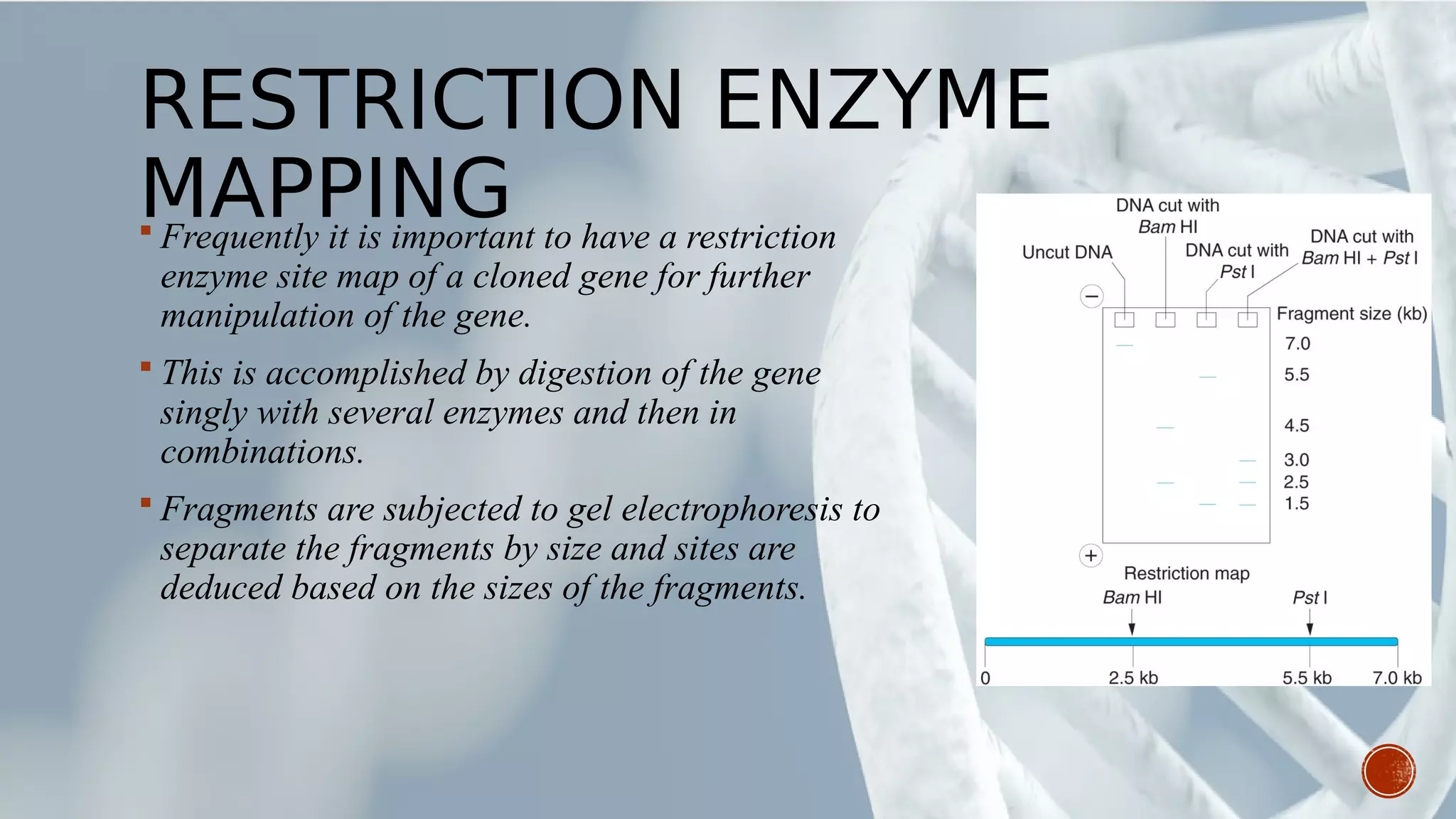
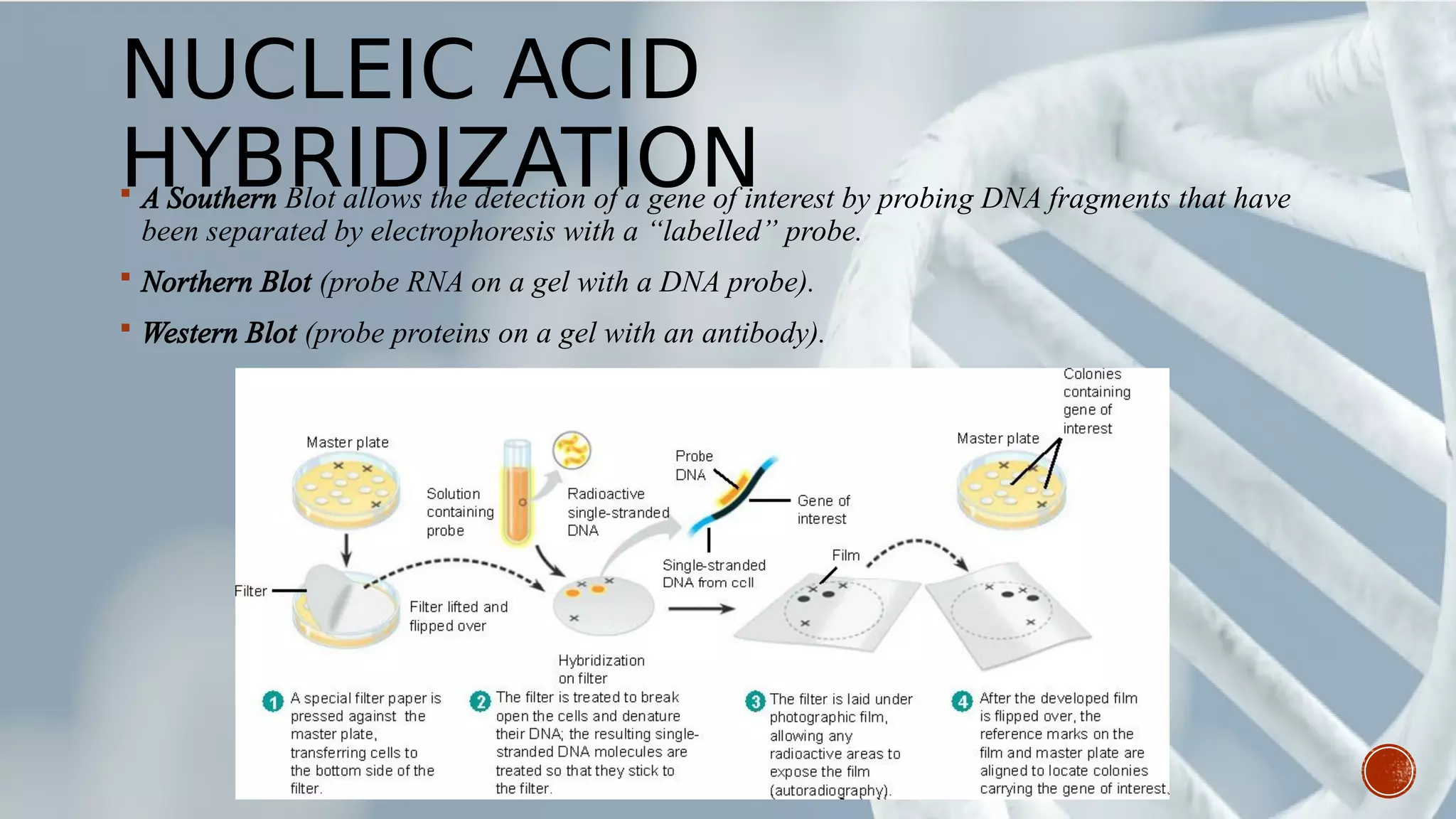
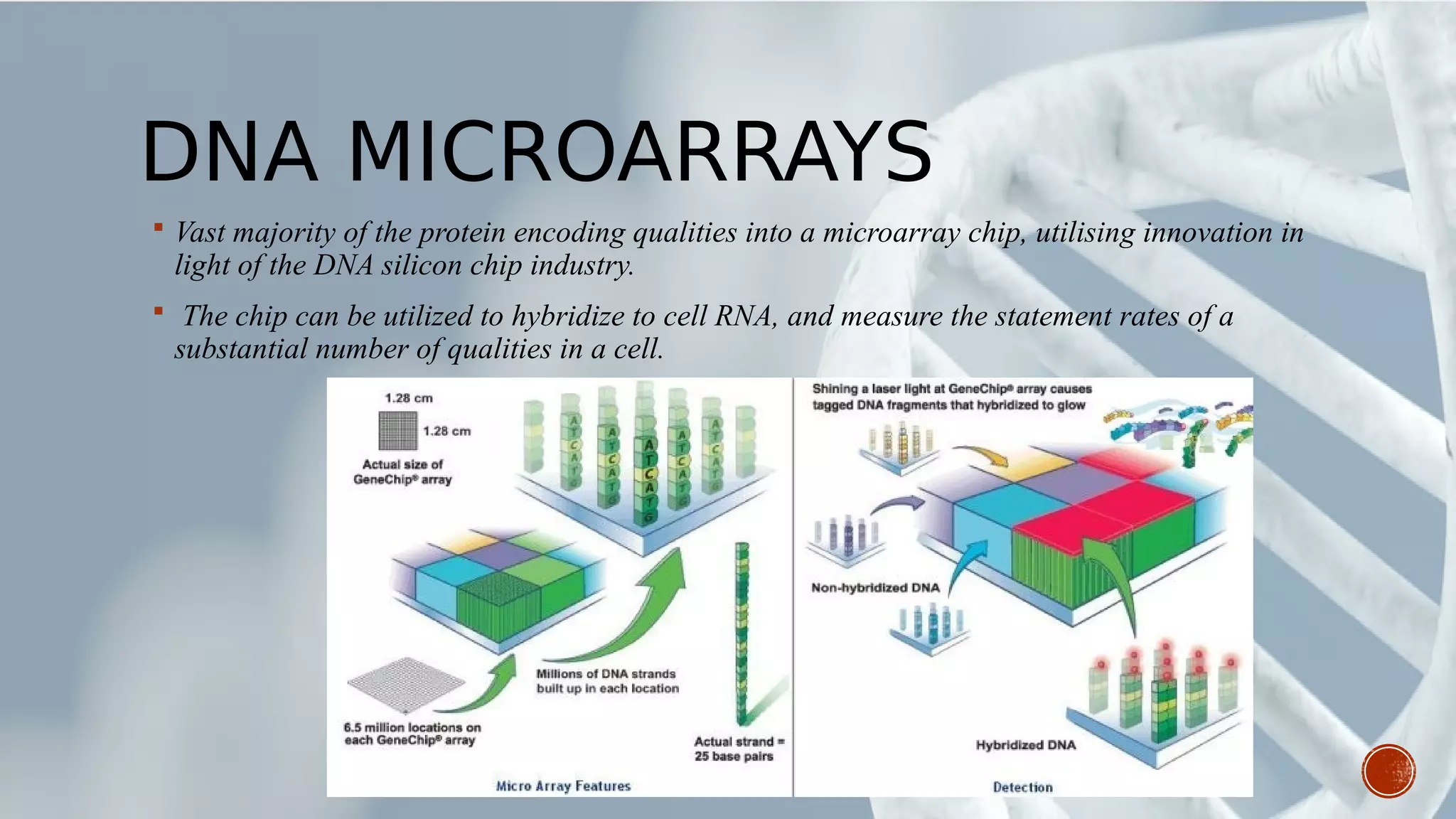
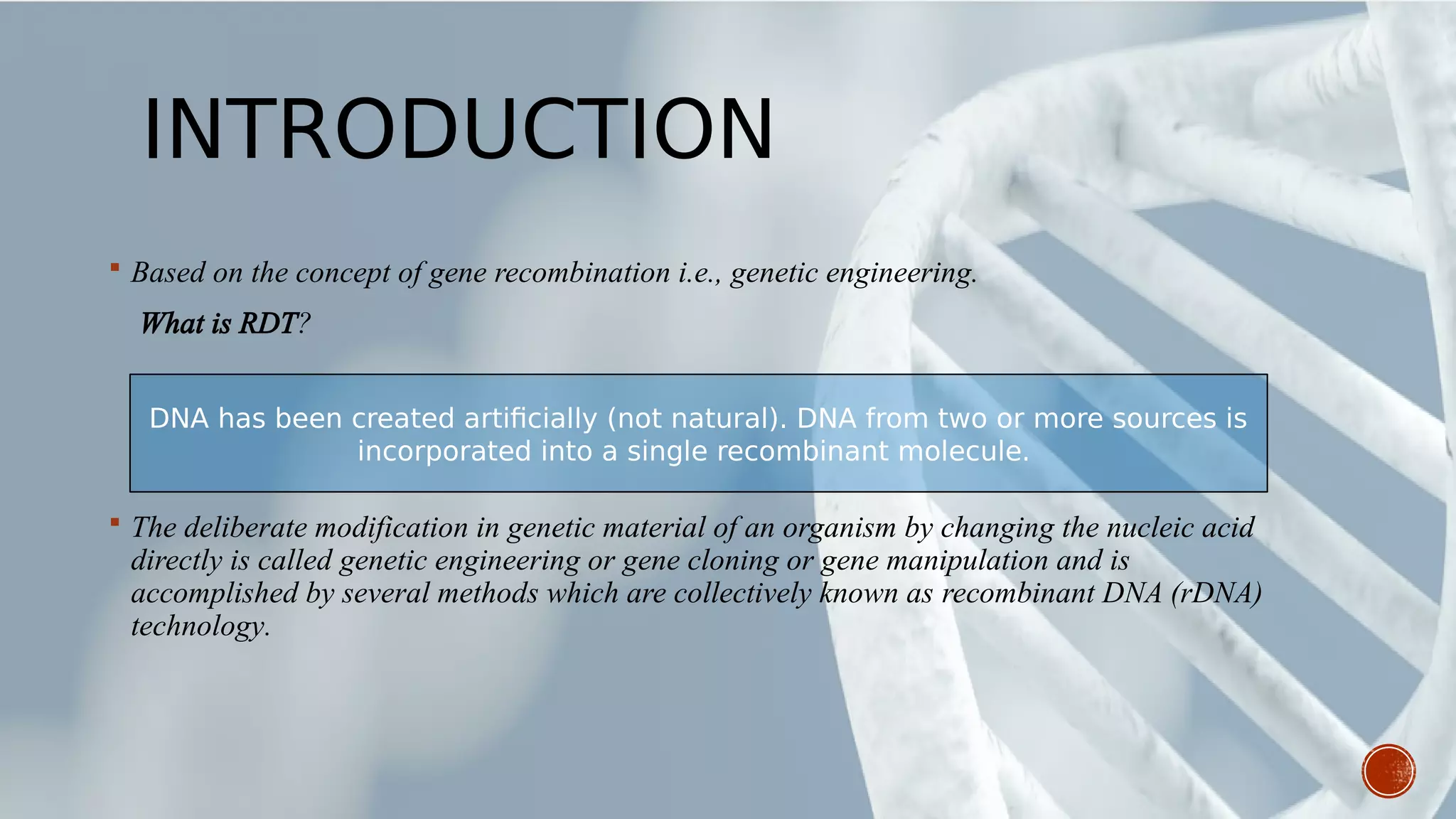
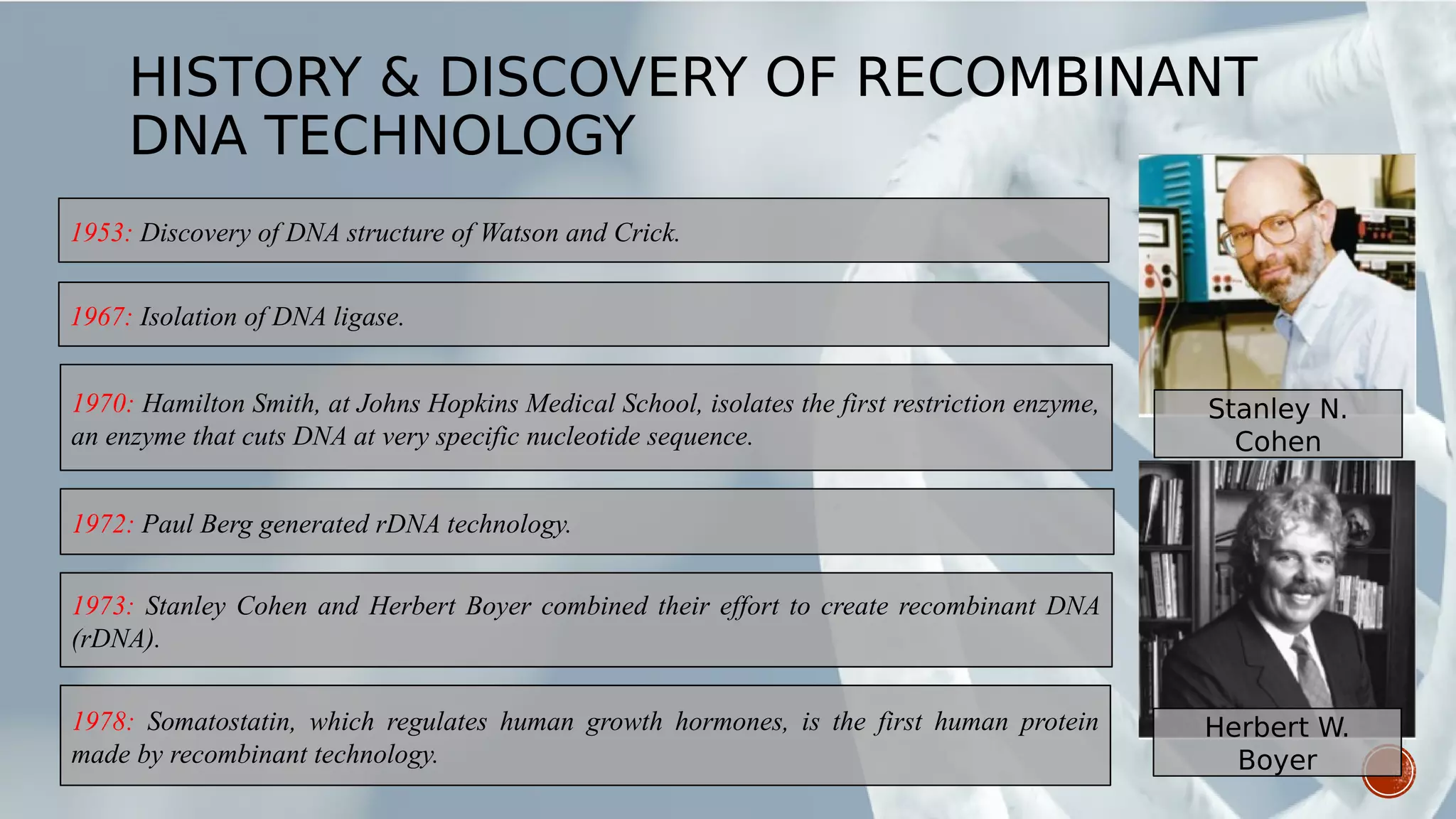
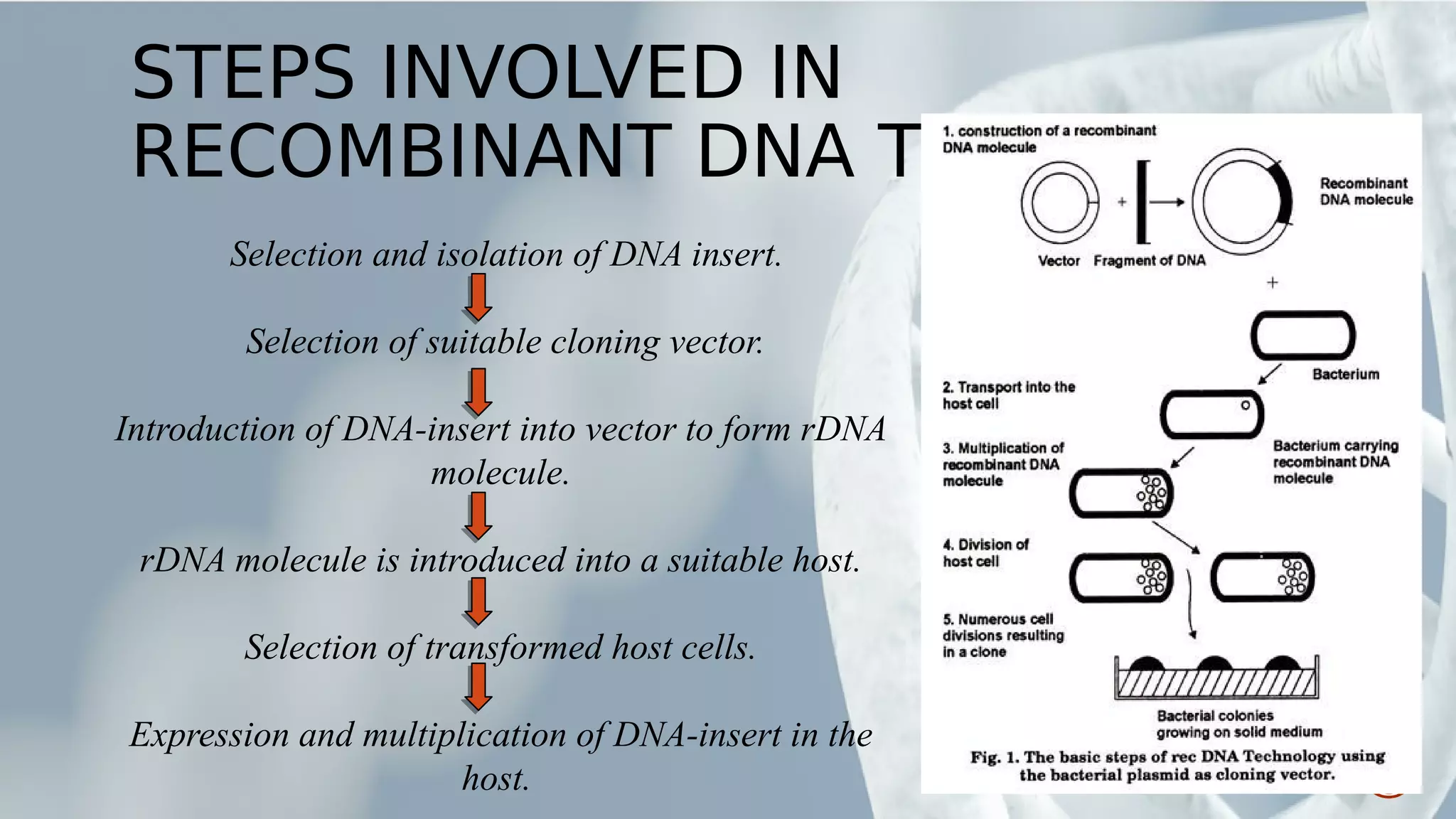
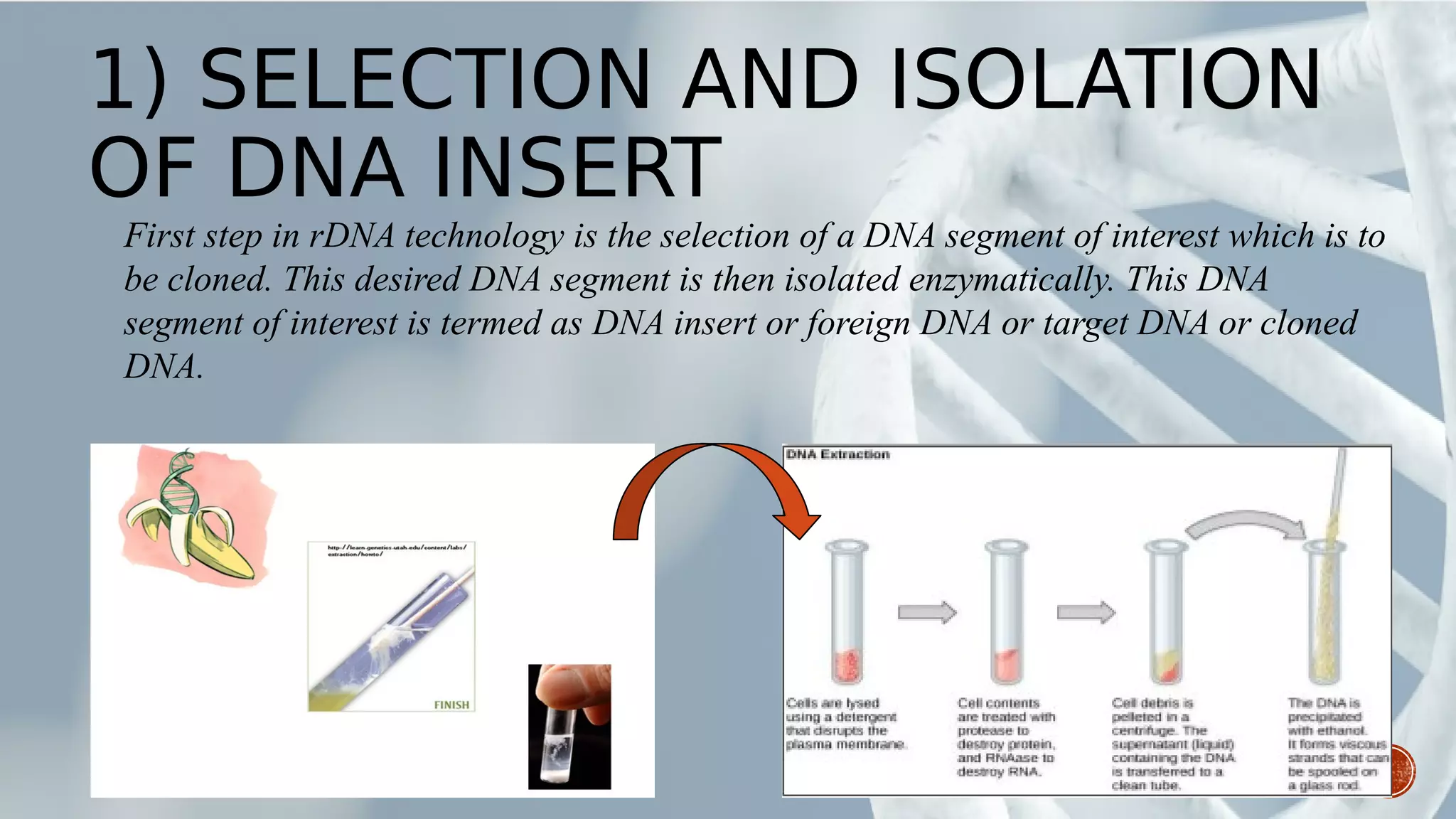
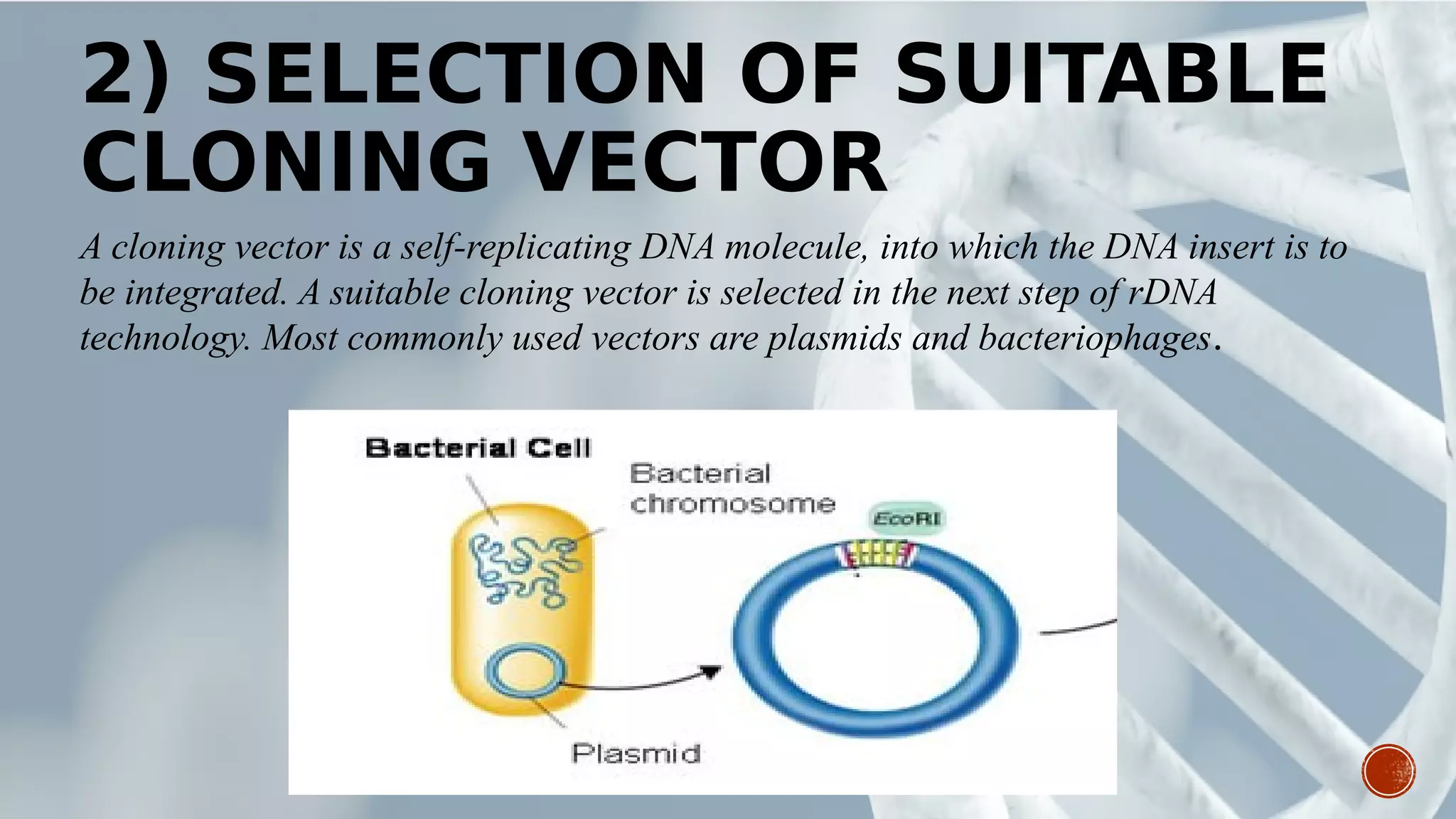
This document provides an overview of recombinant DNA technology (RDT). It discusses the history and discovery of RDT, the key steps involved which include selecting DNA inserts and vectors, introducing the DNA into host cells, and expressing the DNA. Important tools for RDT are described, including restriction enzymes, vectors, and host organisms. Common applications of RDT include producing hormones, medicines, transgenic plants and animals. Ongoing projects utilizing RDT are mentioned.



![3) INTRODUCTION OF DNA-INSERT INTO
VECTOR TO FORM RECOMBINANT DNA
MOLECULE
The target DNA or the DNA insert
which has been extracted and cleaved
enzymatically by the selective
restriction endonuclease enzymes [in
step (1)] are now ligated (joined) by
the enzyme ligase to vector DNA to
form a rDNA molecule which is often
called as cloning-vector-insert DNA
construct.](https://image.slidesharecdn.com/recombinantdnatechnologyrdt-200601080230/75/Recombinant-dna-technology-rdt-9-2048.jpg)
![4) RECOMBINANT DNA MOLECULE IS
INTRODUCED INTO A SUITABLE HOST
Suitable host cells are selected and
the rec DNA molecule so formed [in
step (3)] is introduced into these
host cells. This process of entry of
rec DNA into the host cell is called
transformation. Usually selected
hosts are bacterial cells like E. coli,
however yeast, fungi may also be
utilized.](https://image.slidesharecdn.com/recombinantdnatechnologyrdt-200601080230/75/Recombinant-dna-technology-rdt-10-2048.jpg)