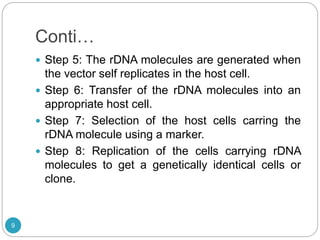
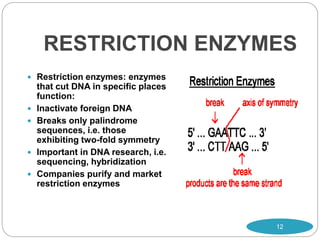
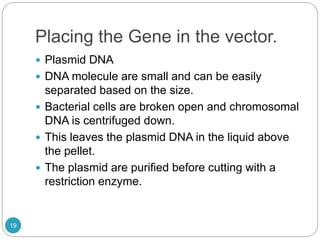
Recombinant DNA technology involves combining DNA molecules from different sources into new combinations that would not otherwise be found in nature. This is done through several steps: isolating the desired DNA fragment, cutting it with restriction enzymes, joining it to a vector molecule like a plasmid, and inserting the new recombinant DNA into a host cell where it can be replicated in large quantities. Applications of this technology include producing human insulin in bacteria for medical use and mapping genomes to study genetic disorders and biological processes. However, some concerns have also been raised about potential negative environmental and safety impacts.