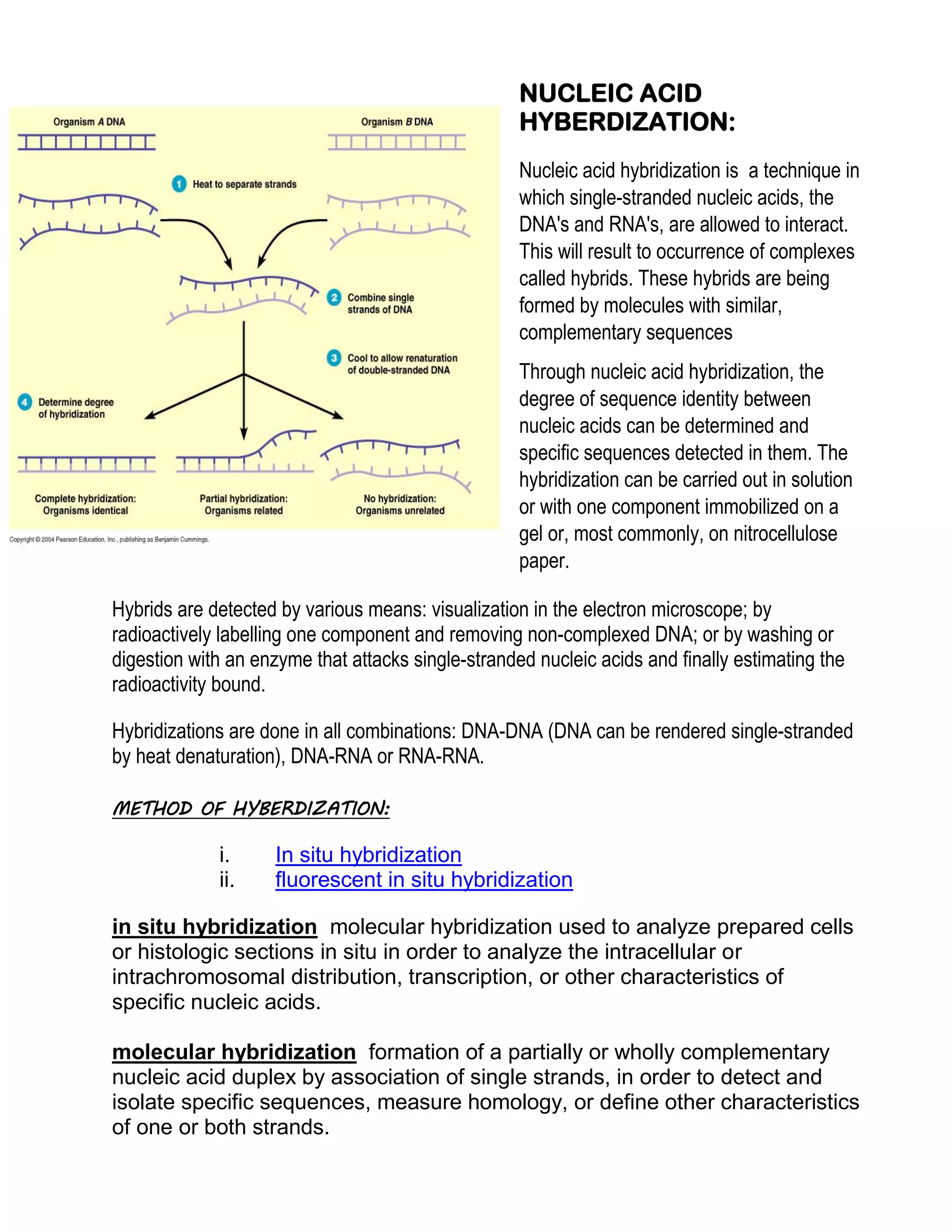
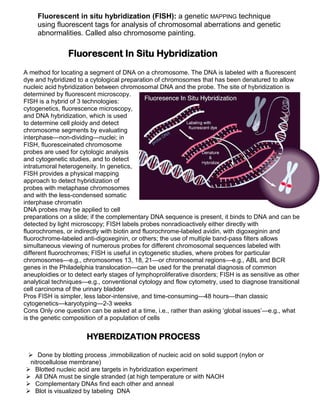
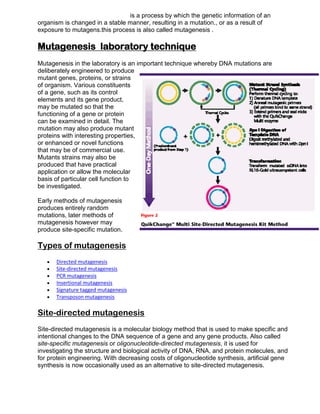
Nucleic acid hybridization is a technique to detect specific nucleic acid sequences. It involves allowing single-stranded DNA or RNA molecules to interact and form hybrids based on complementary base pairing. This allows researchers to determine the degree of sequence identity between nucleic acids and detect specific sequences. Fluorescent in situ hybridization (FISH) is a type of hybridization that uses fluorescent probes to map DNA sequences onto chromosomes. FISH has applications in cytogenetics, cancer detection, and prenatal diagnosis of genetic disorders.