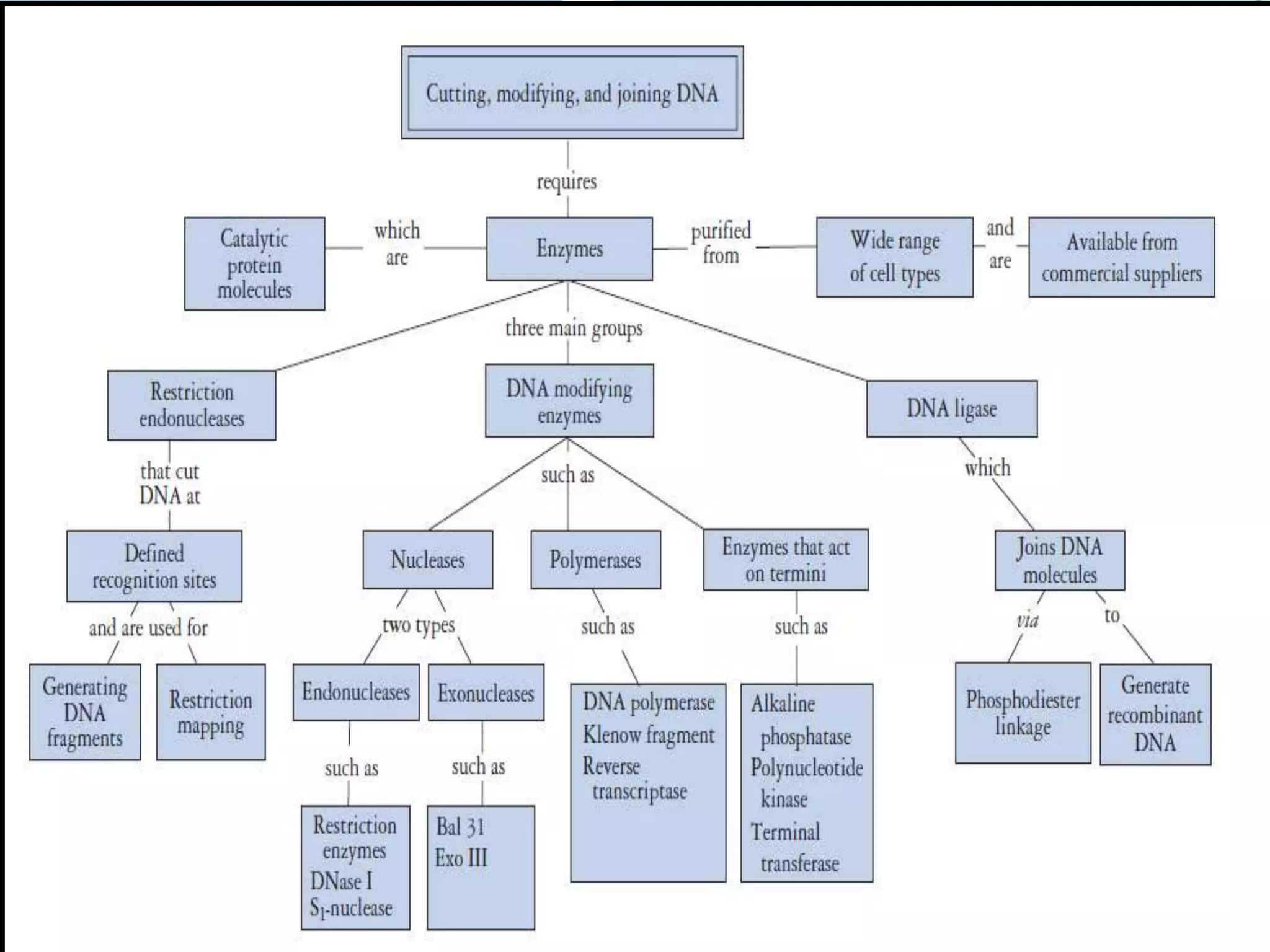
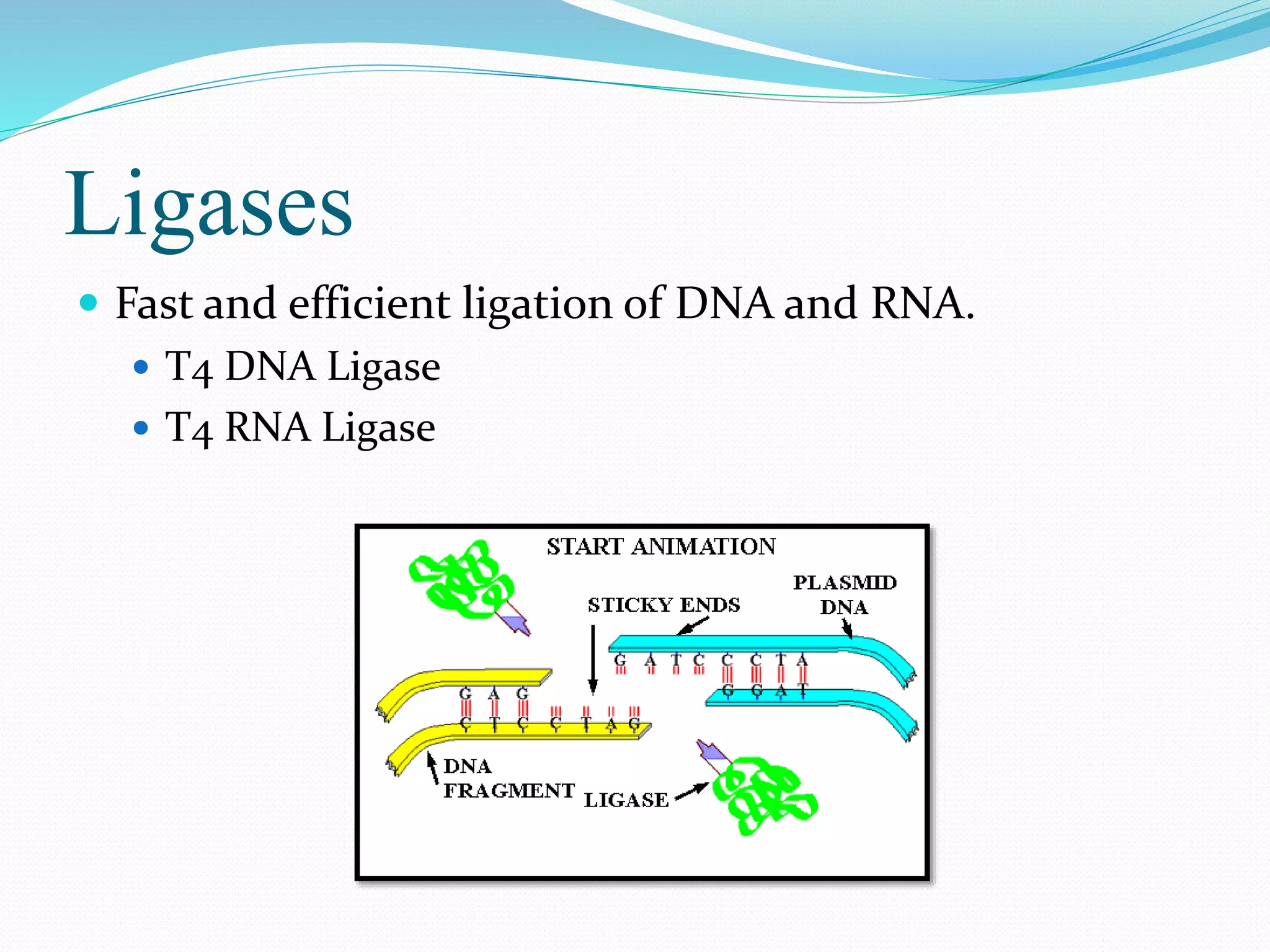
This document discusses various enzymes used for genetic engineering and DNA manipulation. It describes restriction endonucleases and DNA ligase which cut and join DNA fragments. It also discusses other DNA modifying enzymes like nucleases which degrade DNA, and polymerases which synthesize DNA copies. Specific enzymes covered in detail include DNA polymerase I, T4 DNA polymerase, T7 DNA polymerase, terminal transferase, T4 DNA ligase, and T4 RNA ligase.