Embed presentation



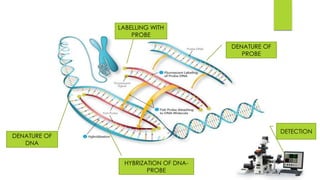

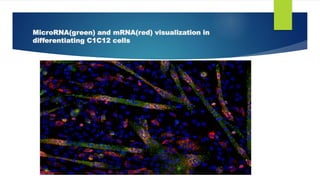






The document details the fluorescent in-situ hybridization (FISH) technique used in cytogenetics, developed in the early 1980s, which employs fluorescent probes to identify specific DNA and RNA sequences in chromosomes. Protocols for FISH include preparation of probes, hybridization, and visualization under a microscope, with applications in genetic counseling, disease diagnosis, and evolutionary studies. FISH is particularly useful for detecting specific genetic conditions and evaluating gene presence or absence across different species.











