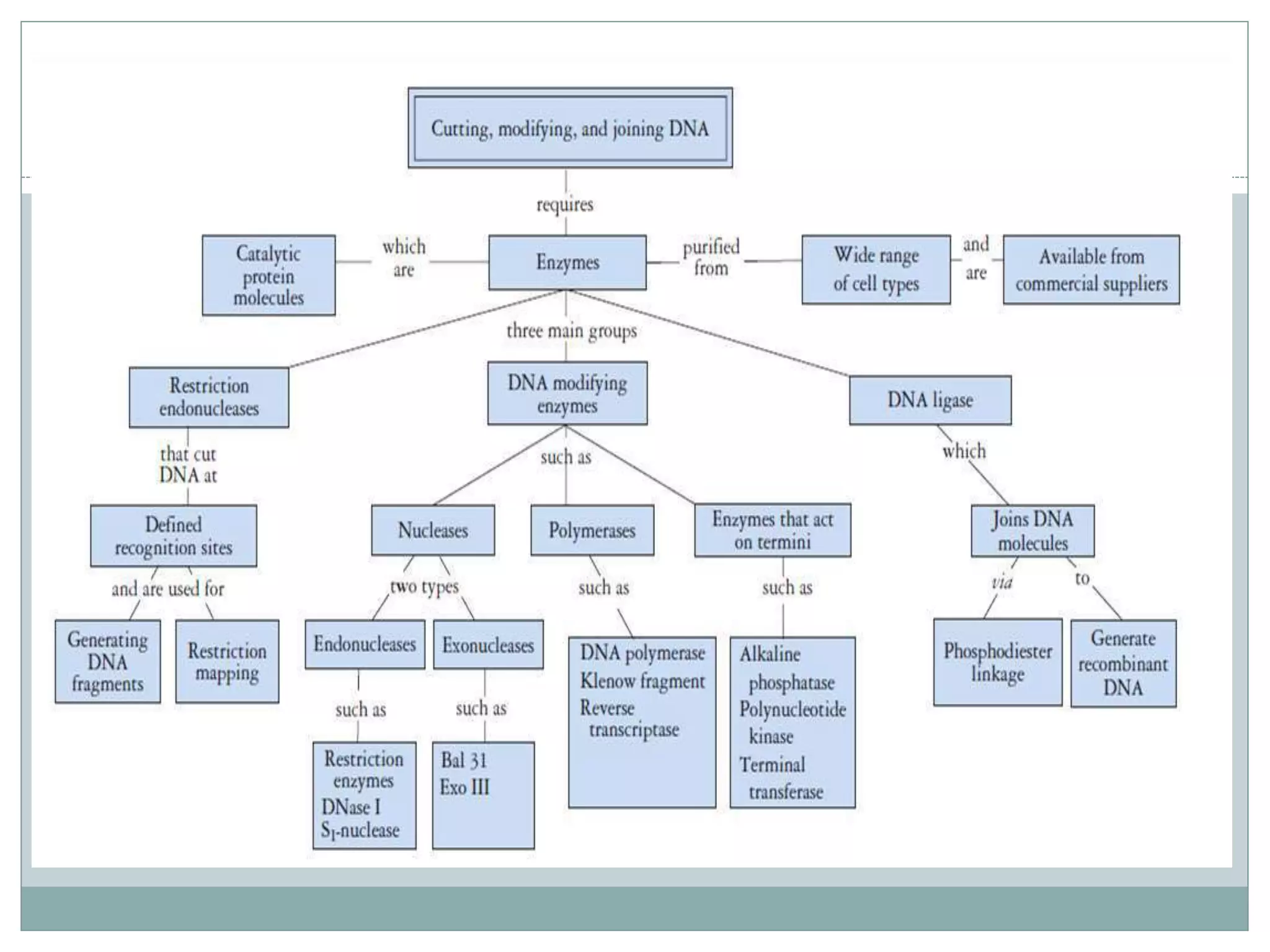
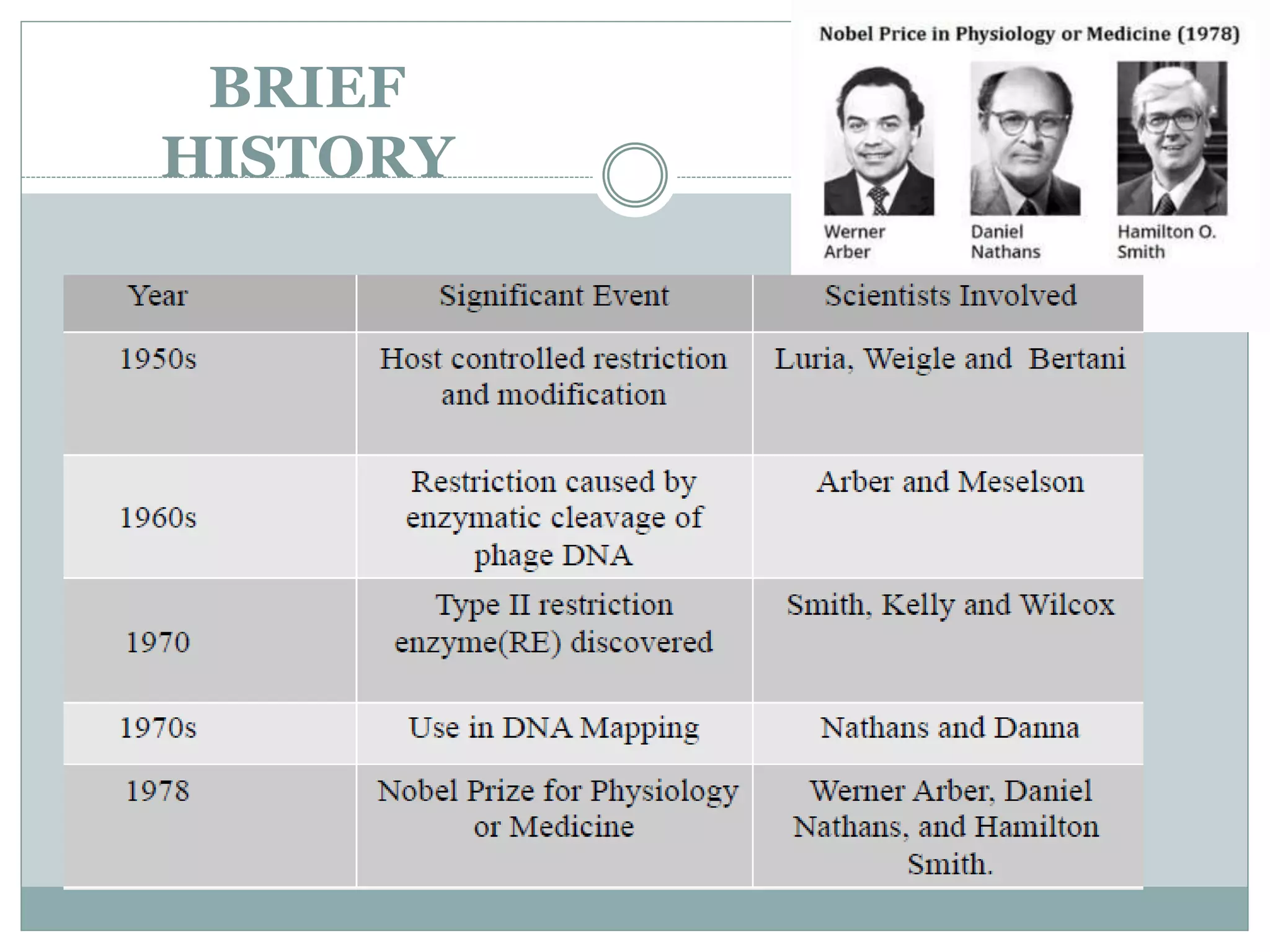
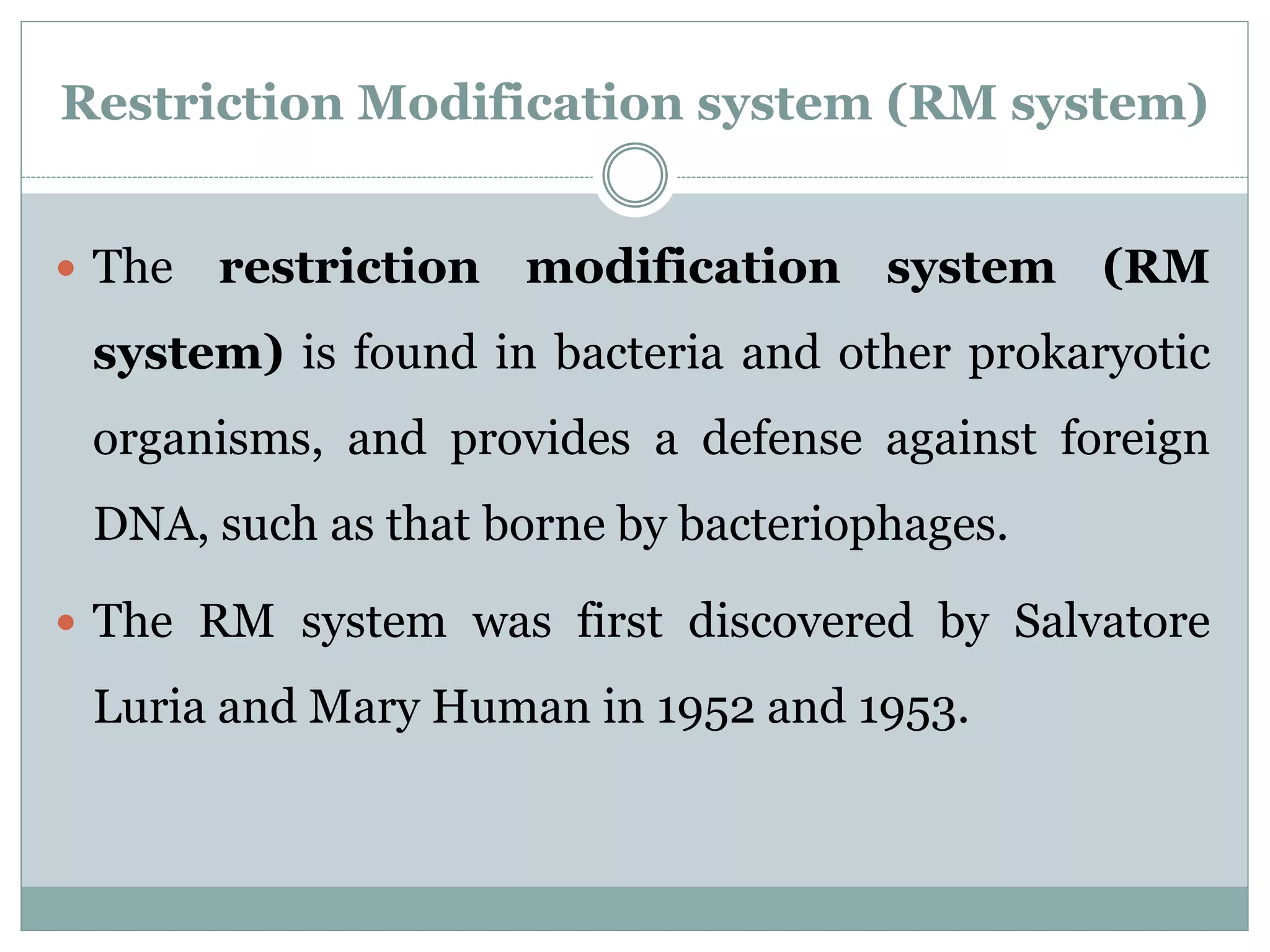
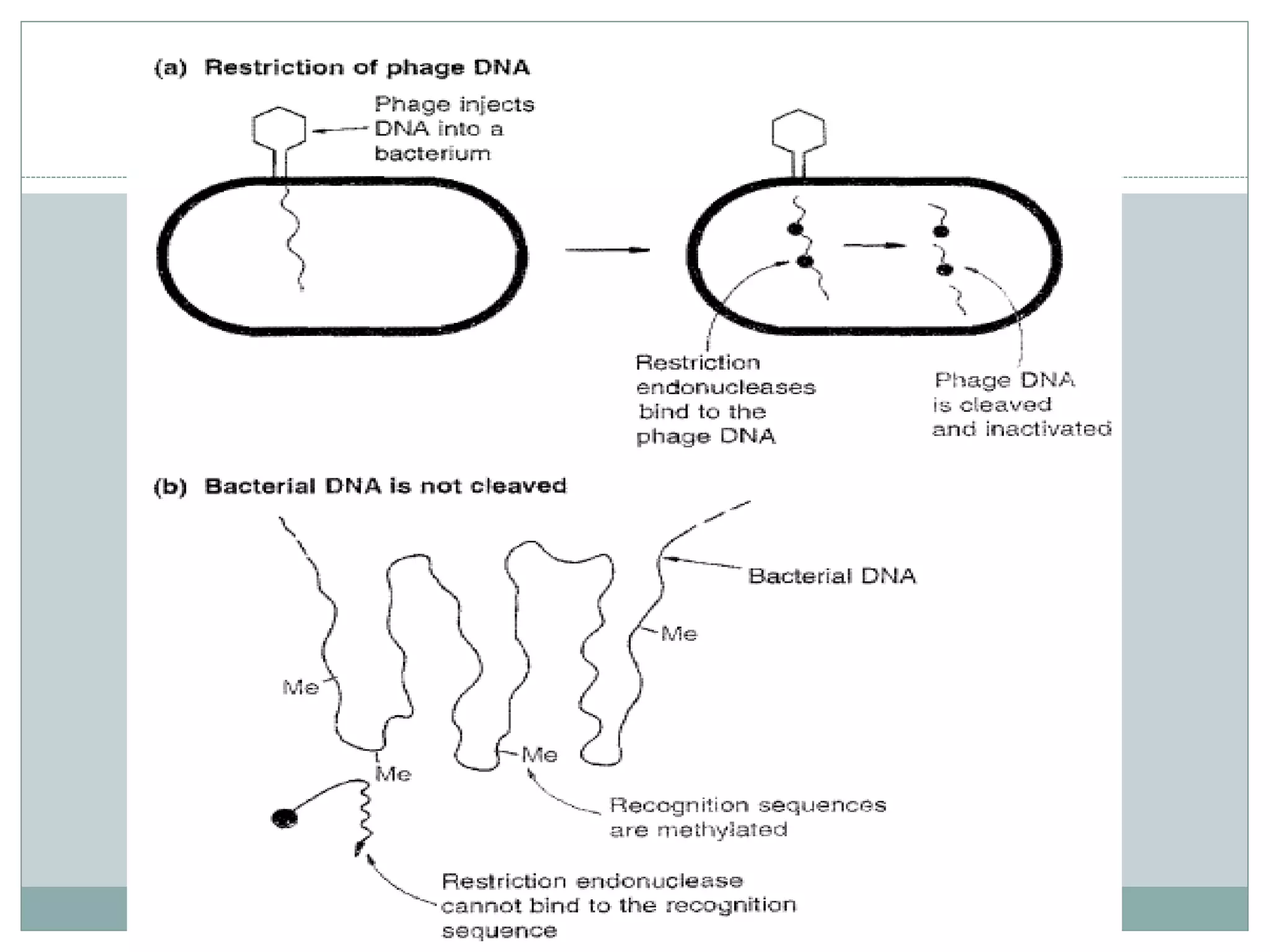
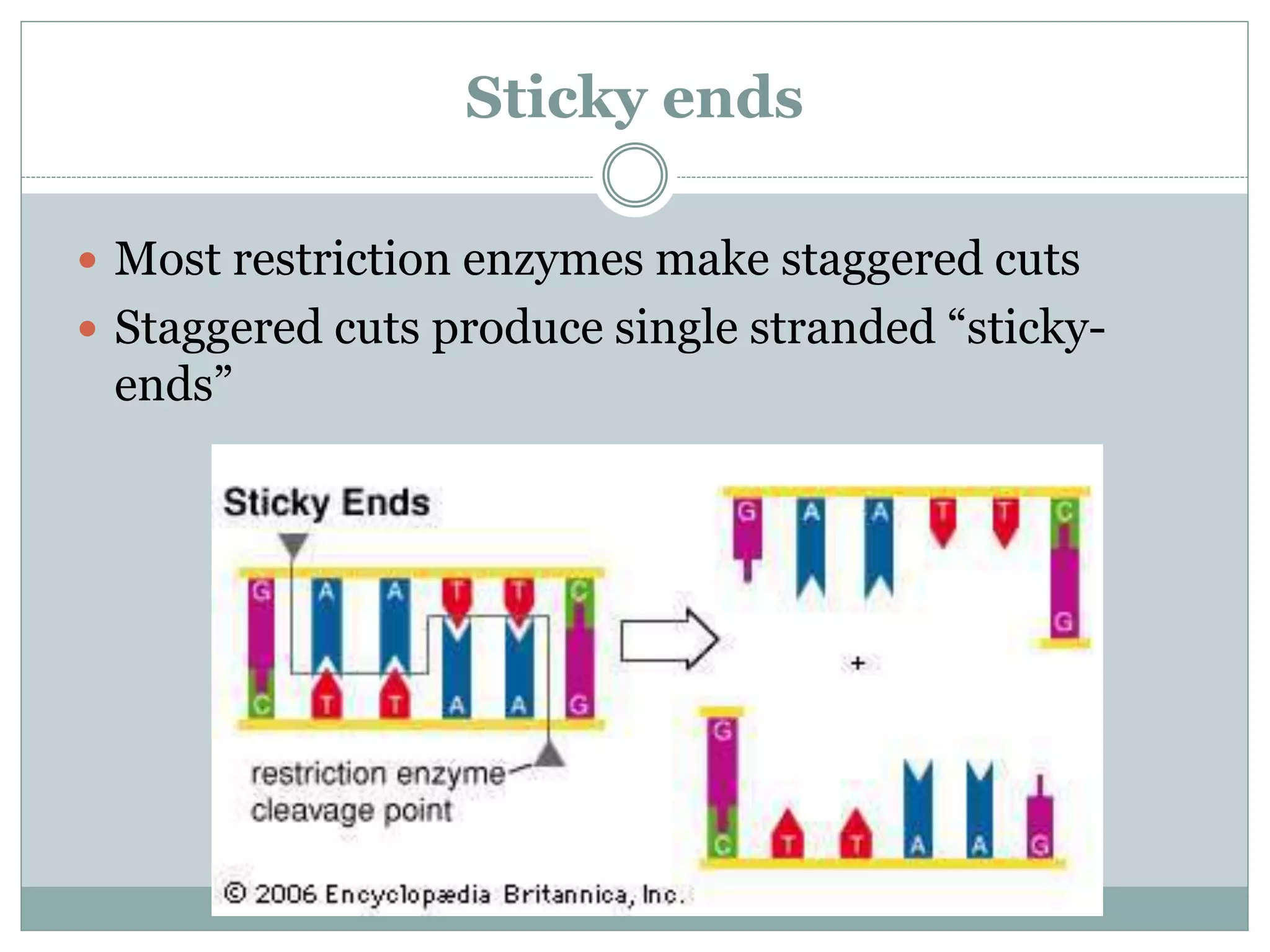
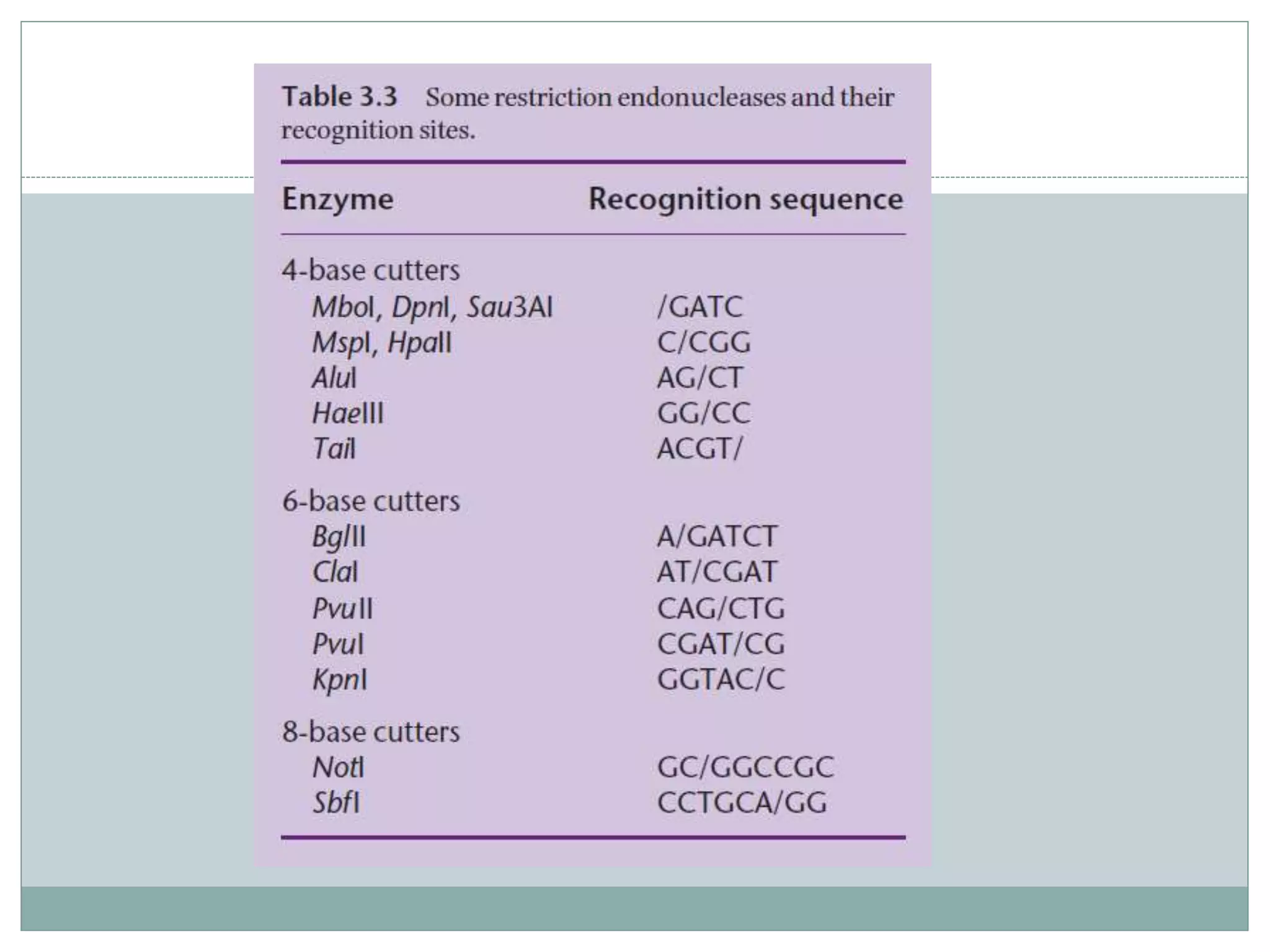
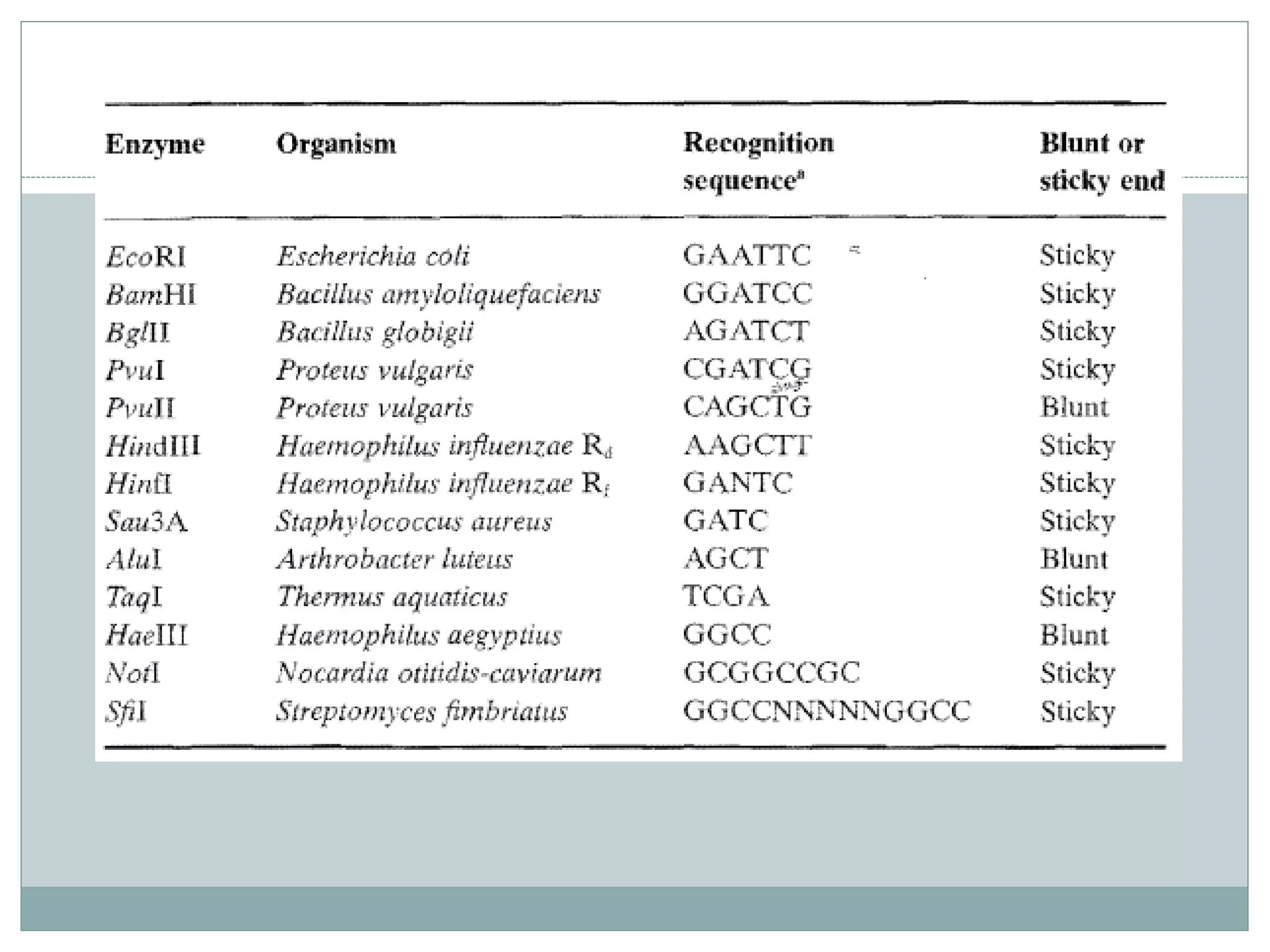
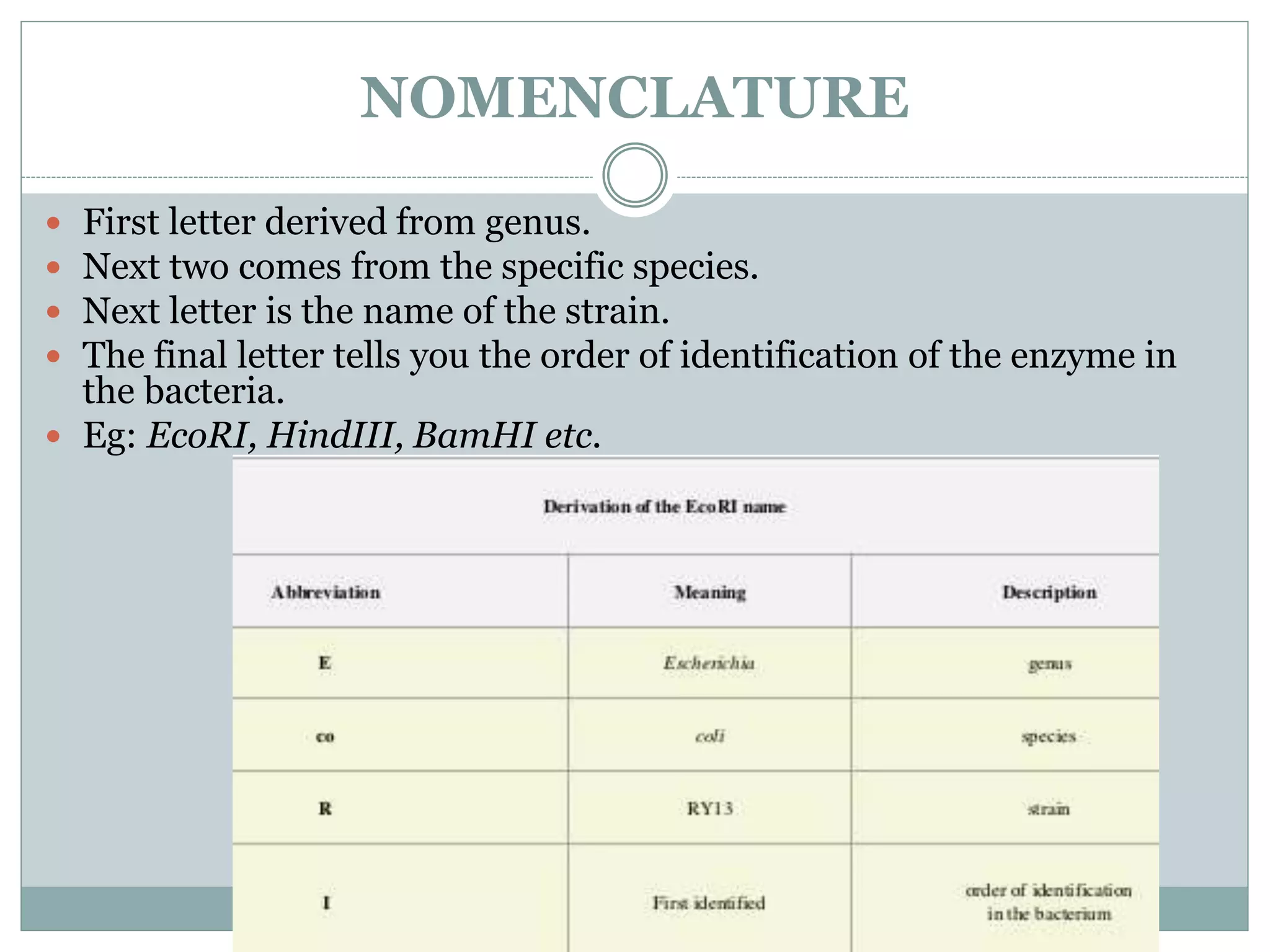
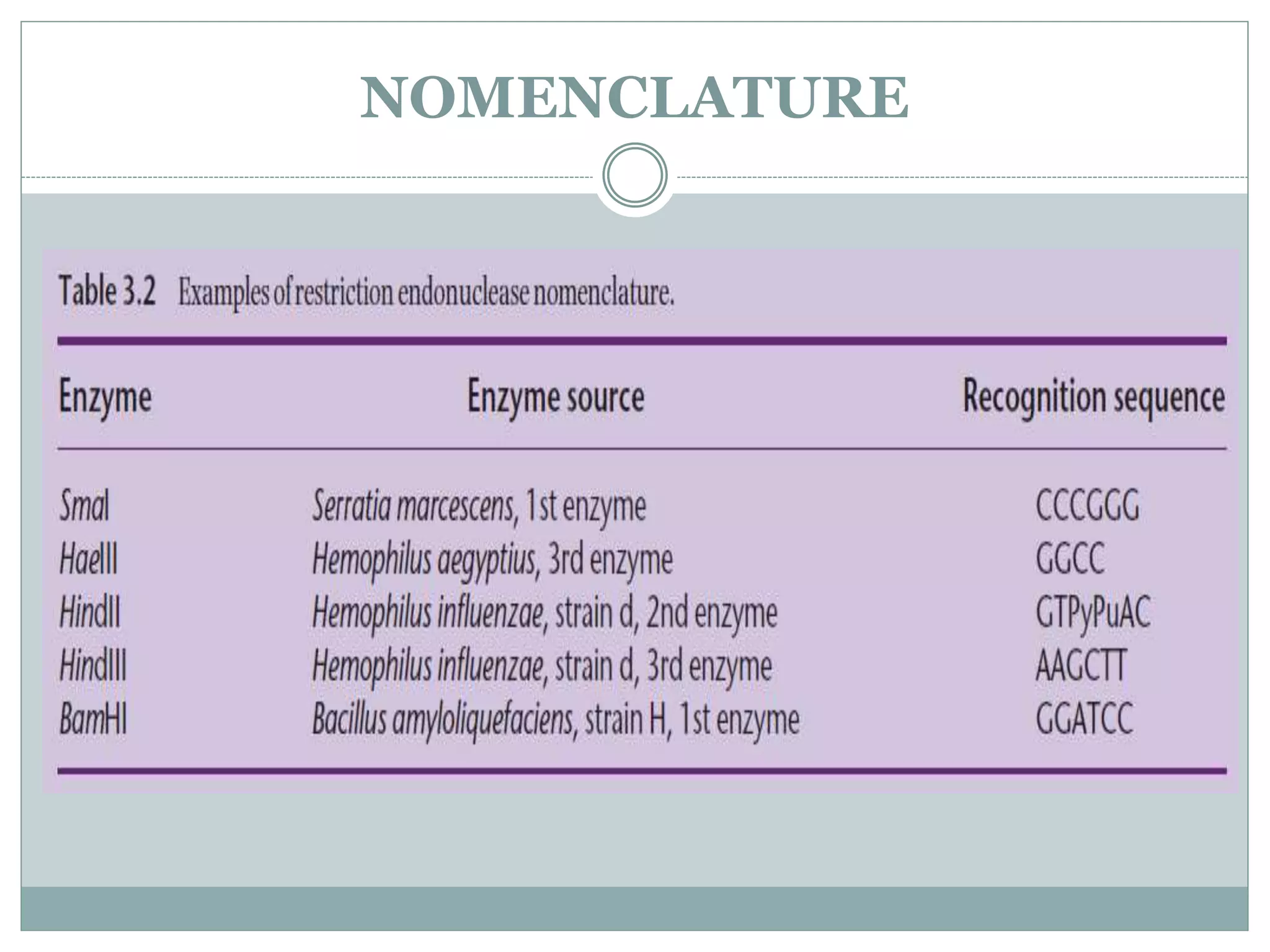
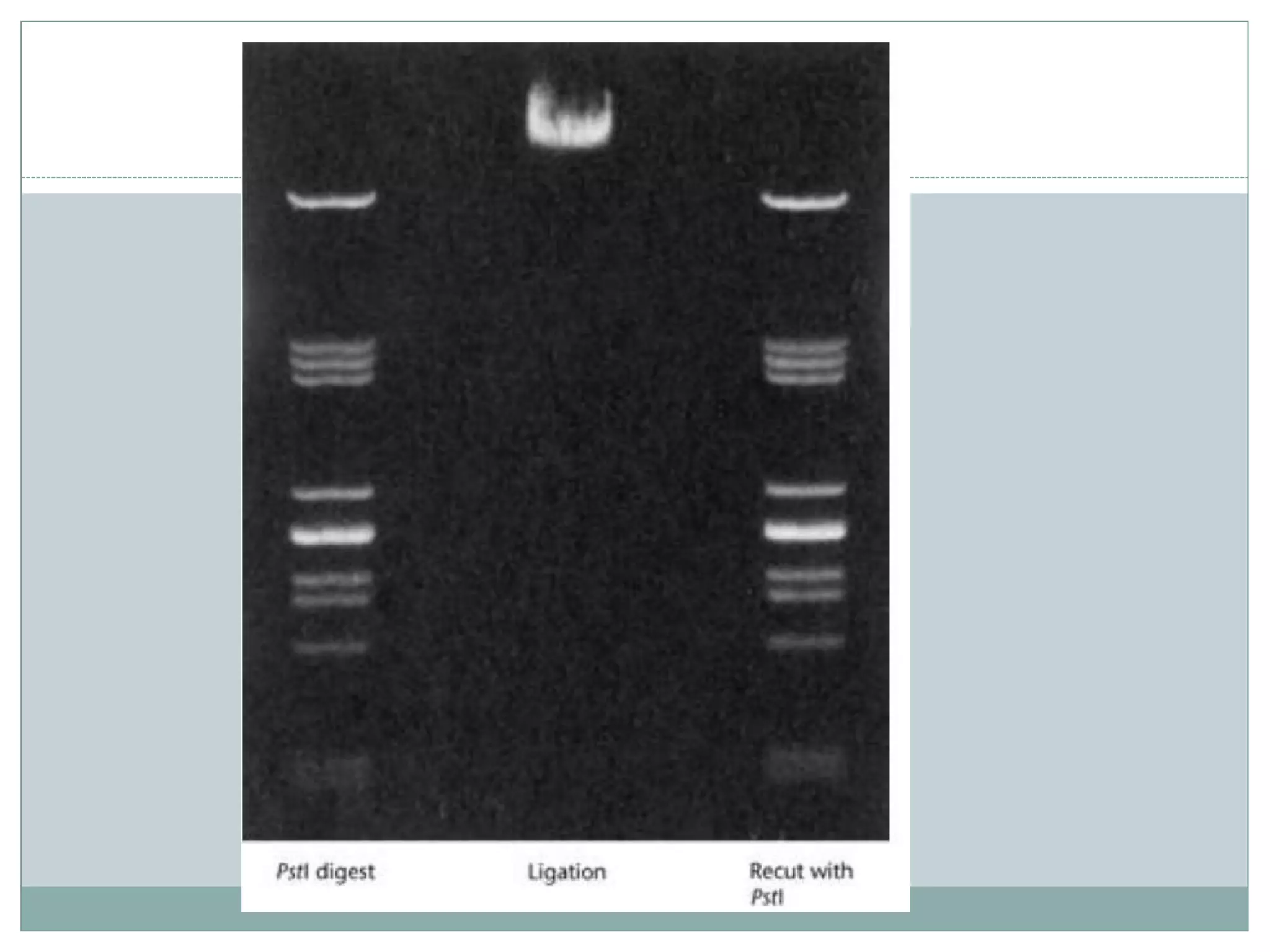
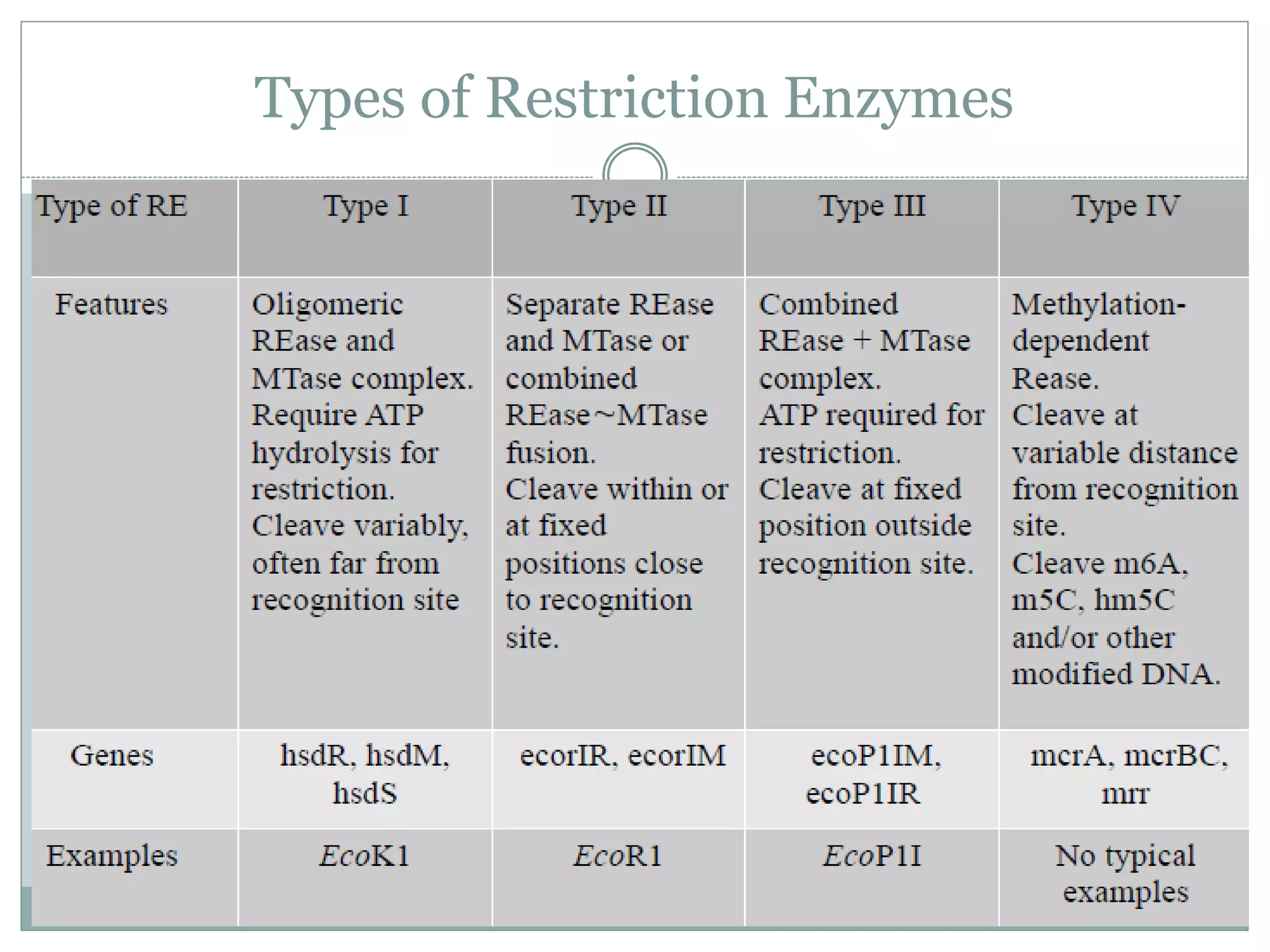
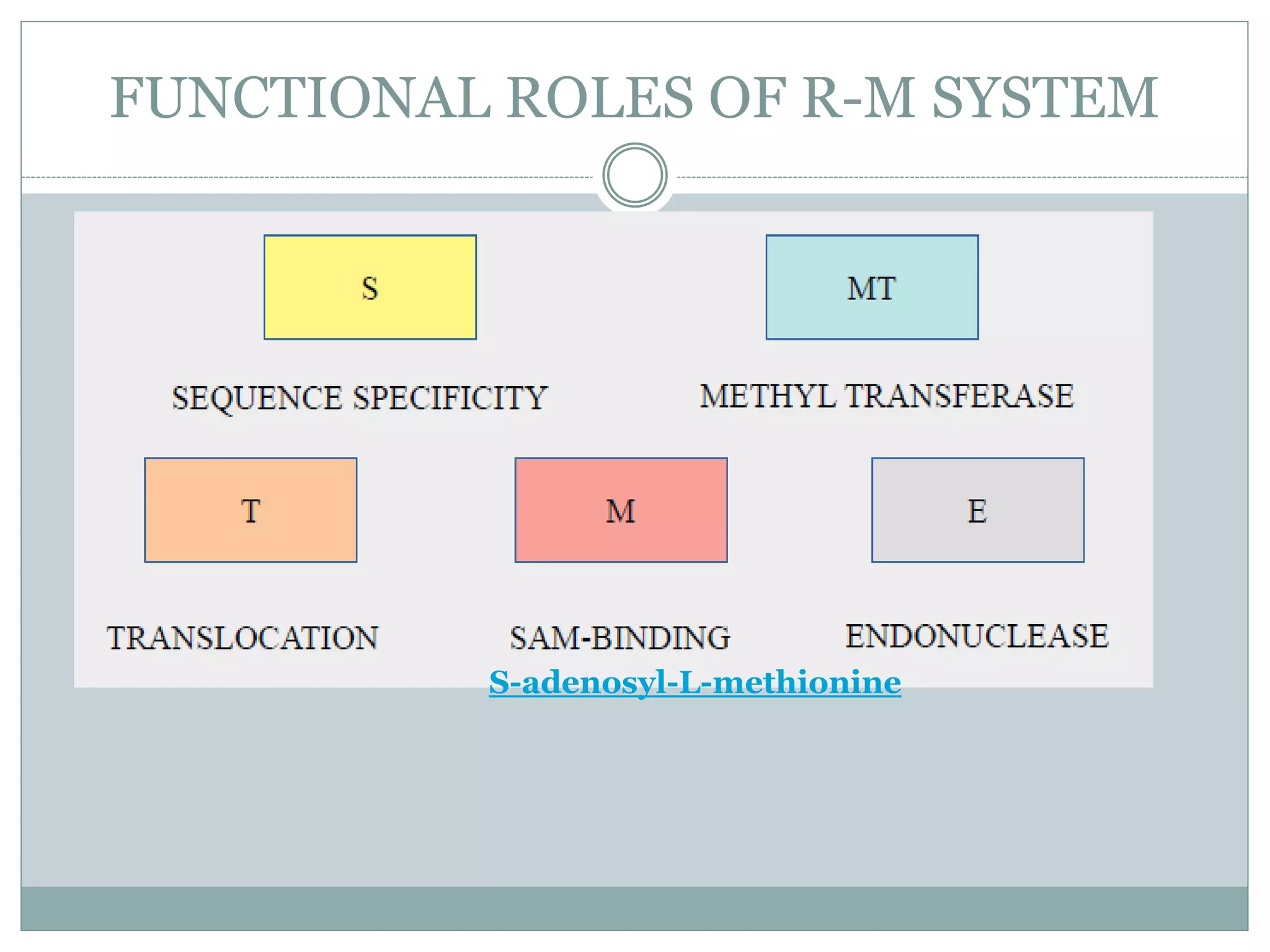
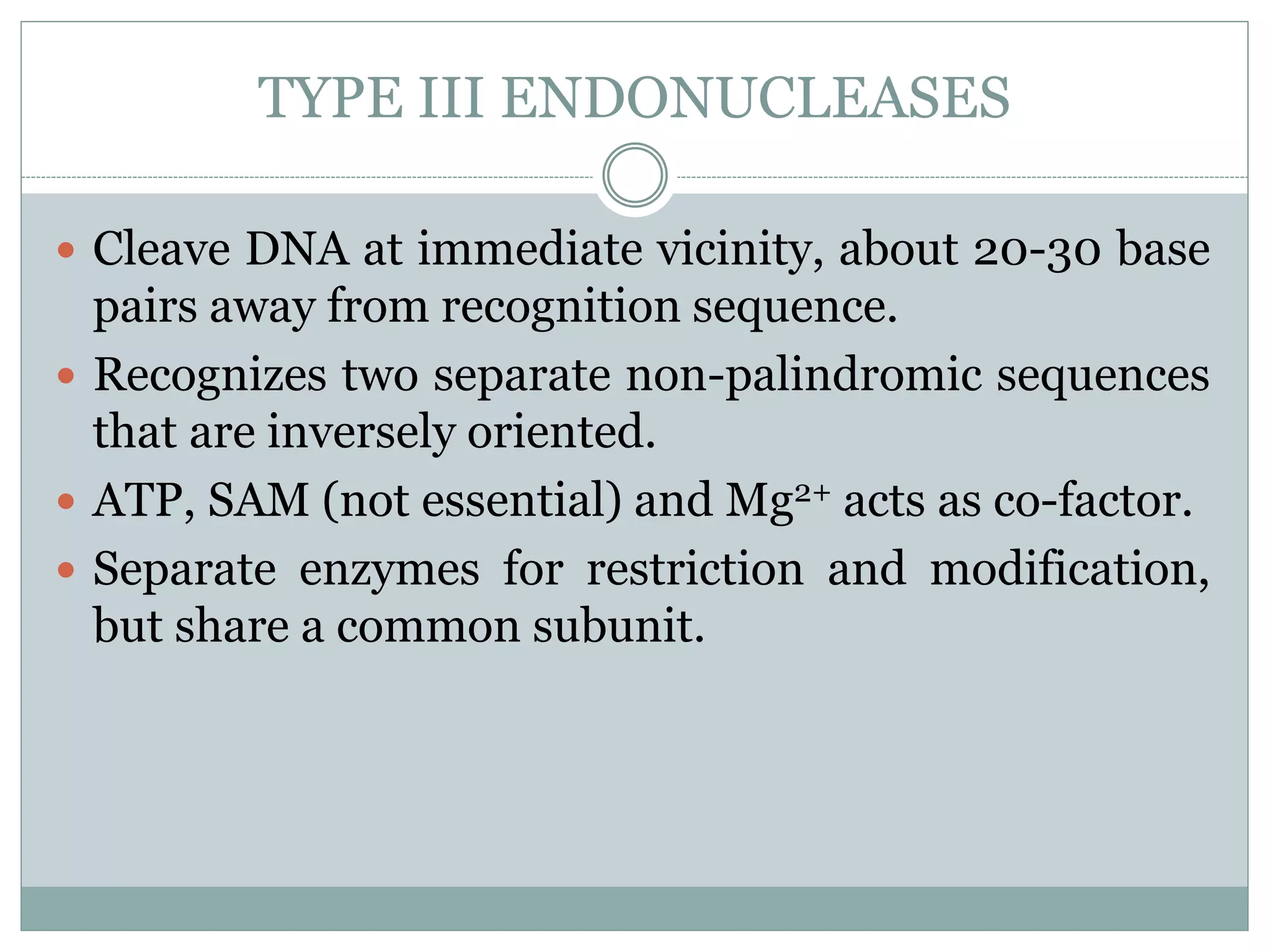
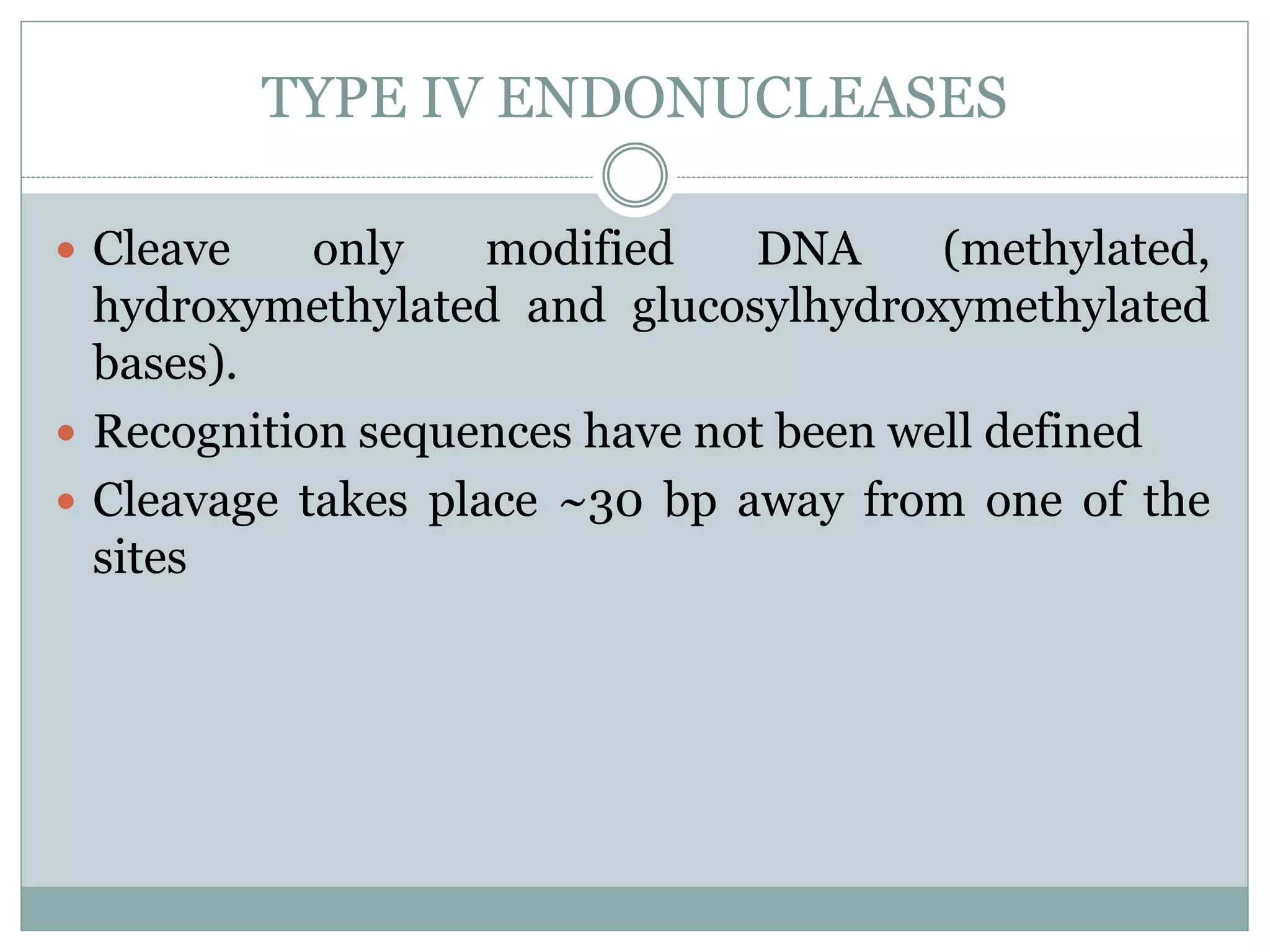
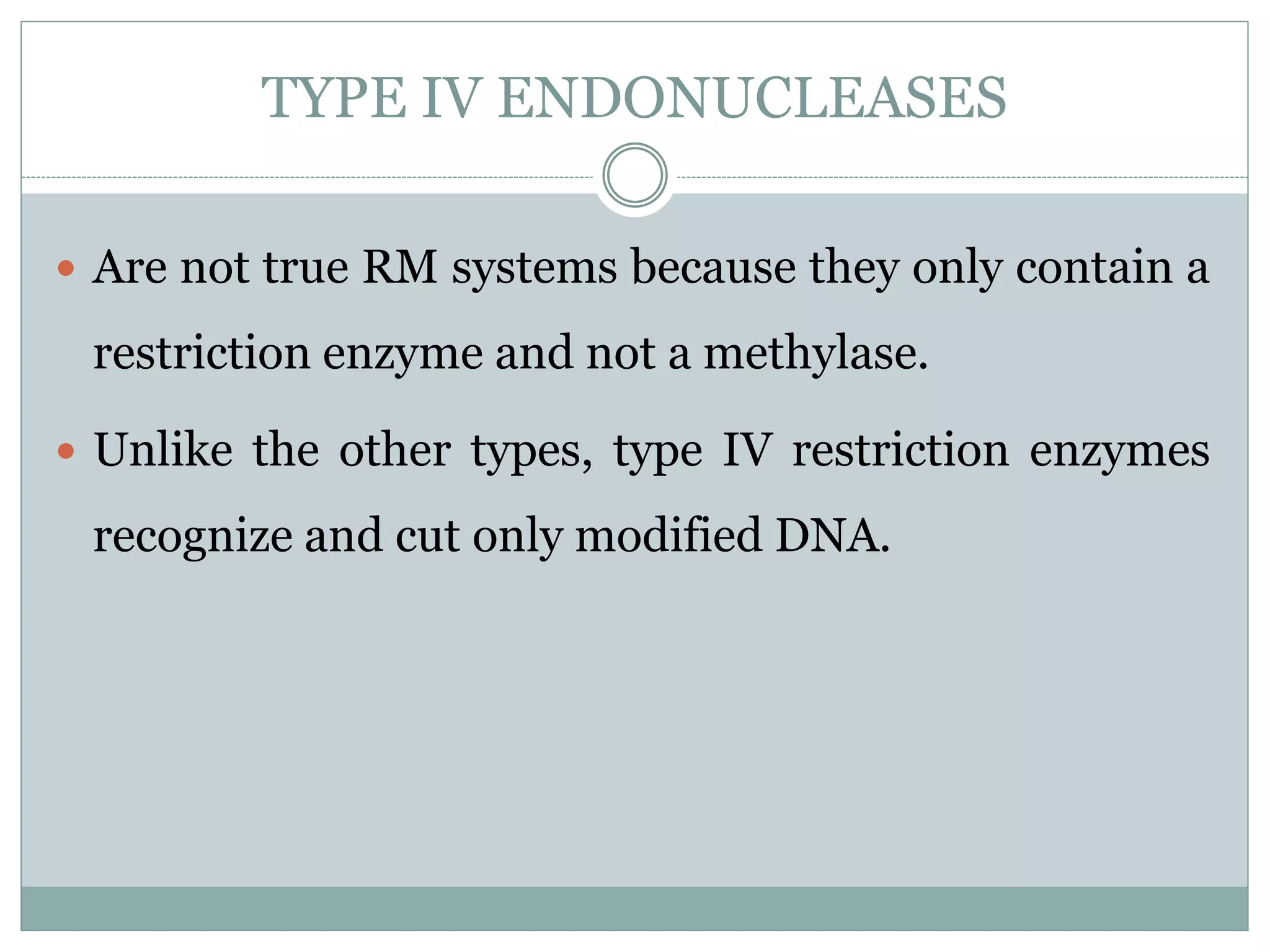
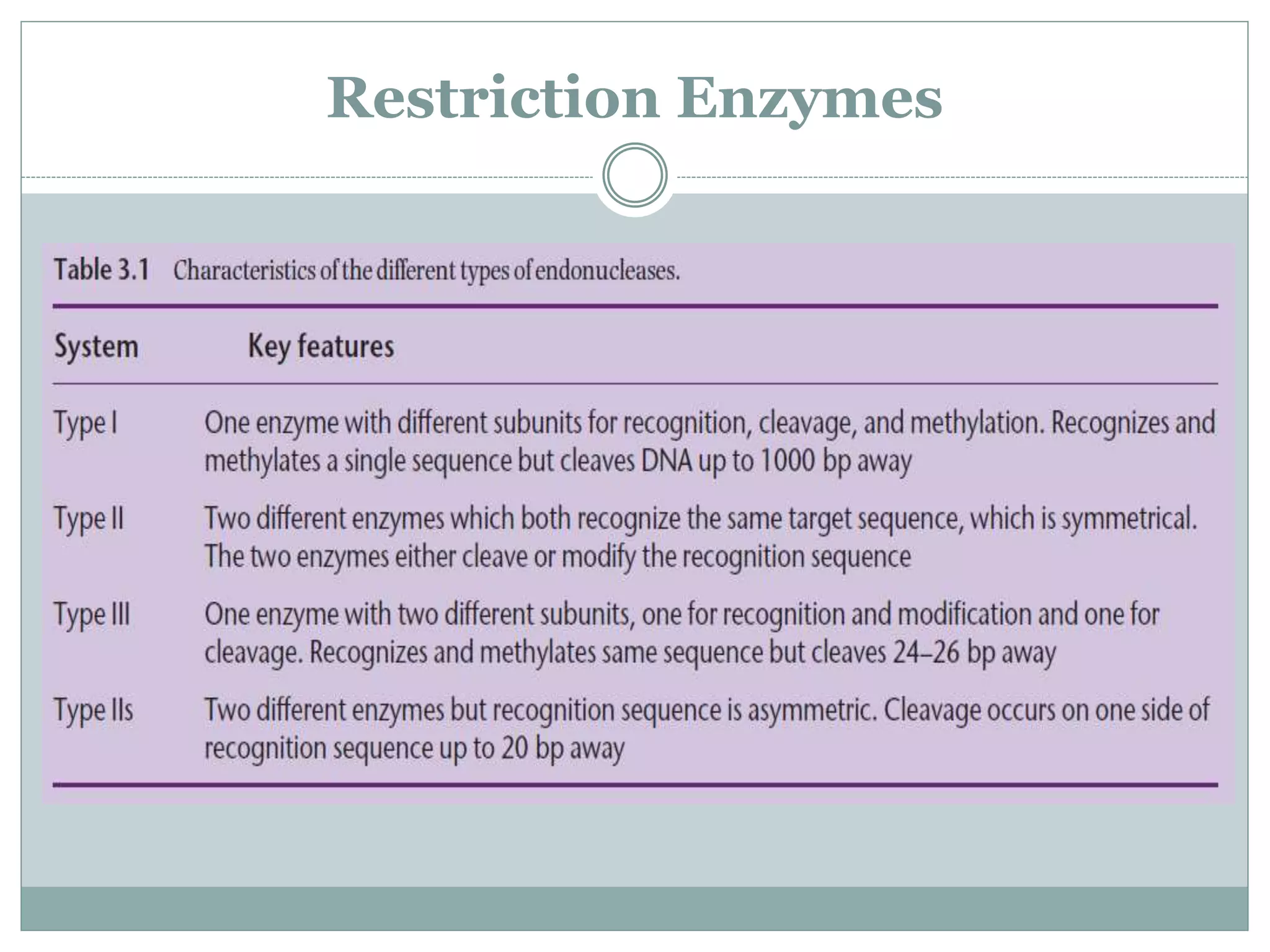
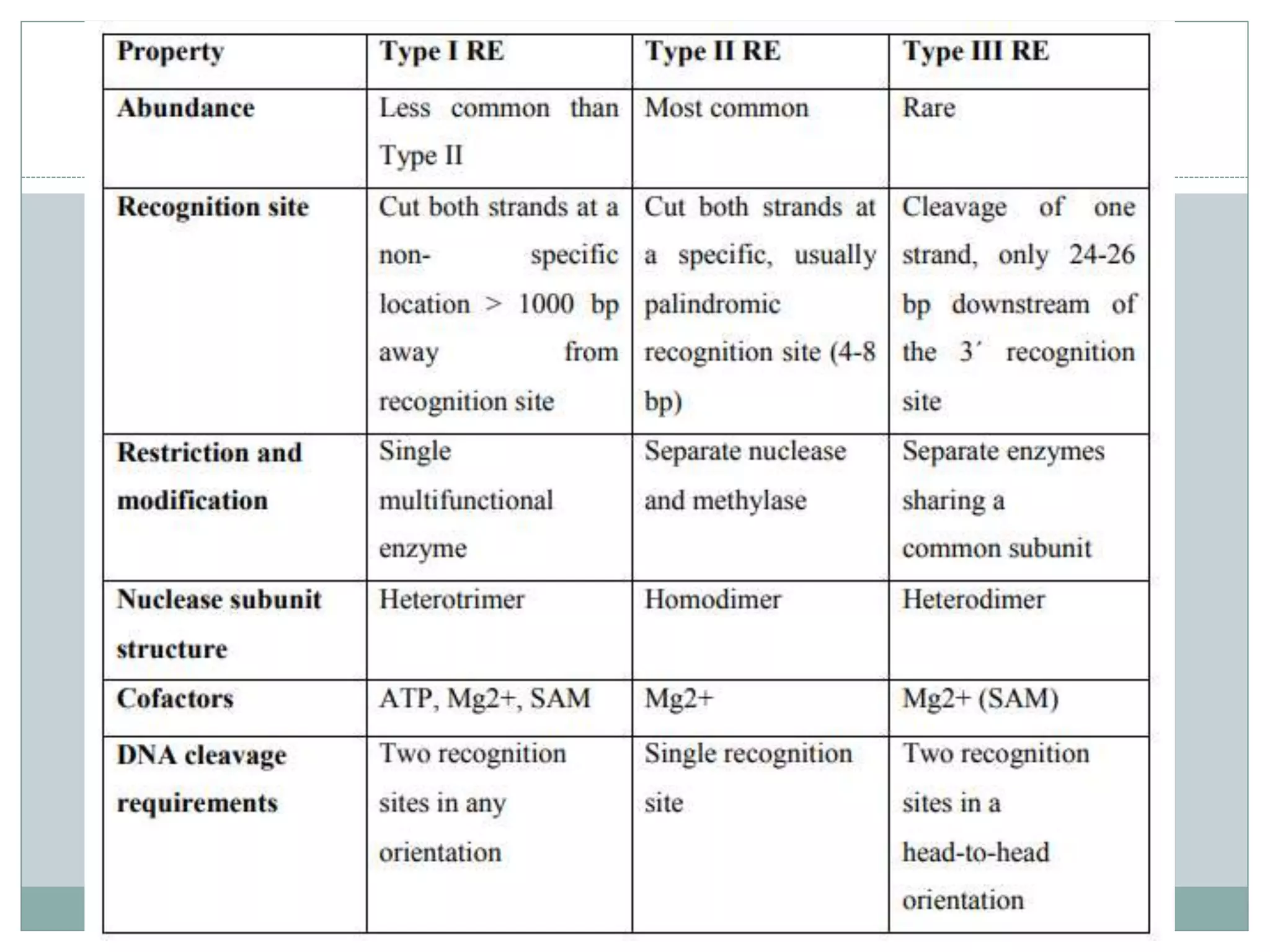
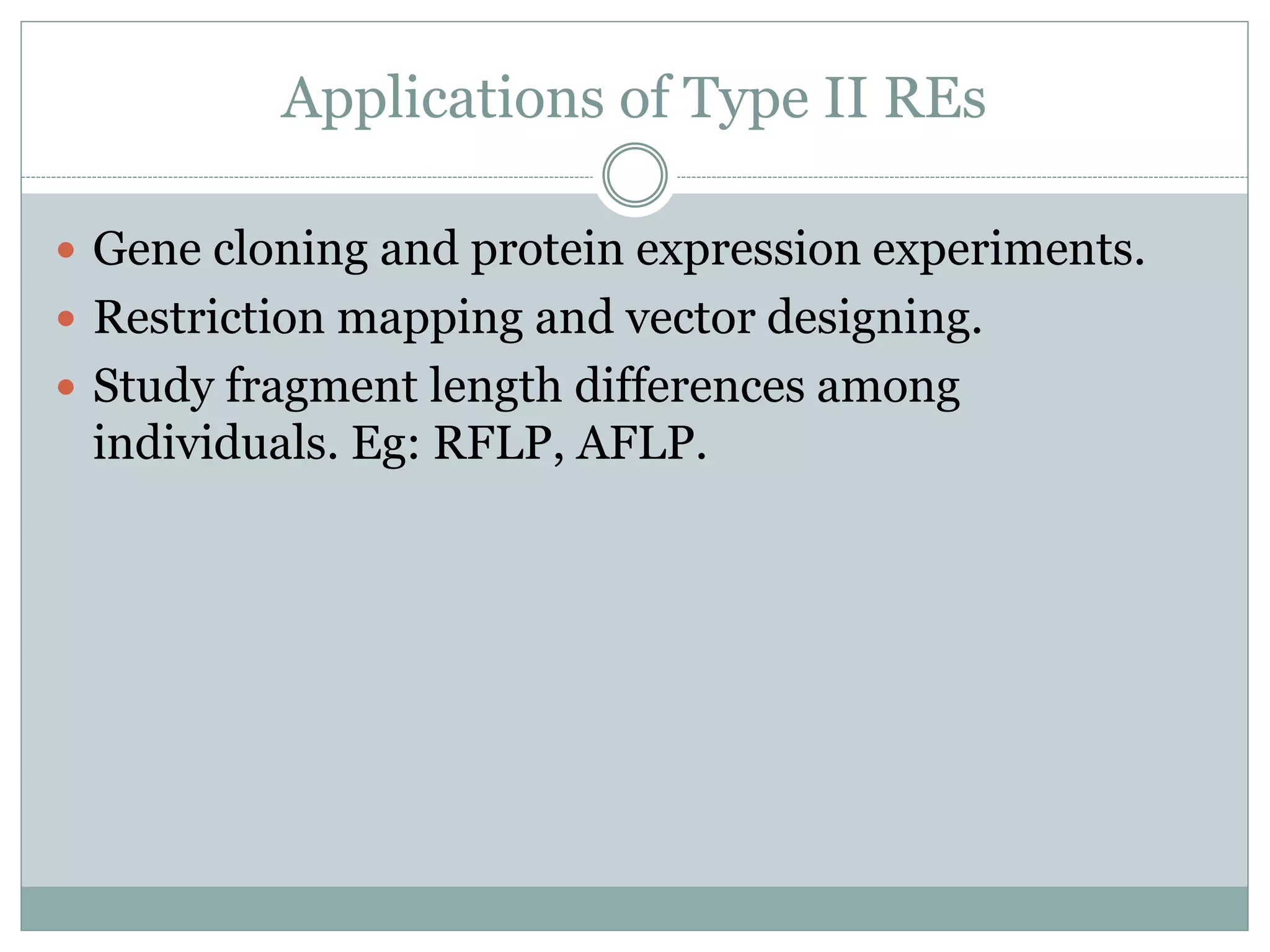
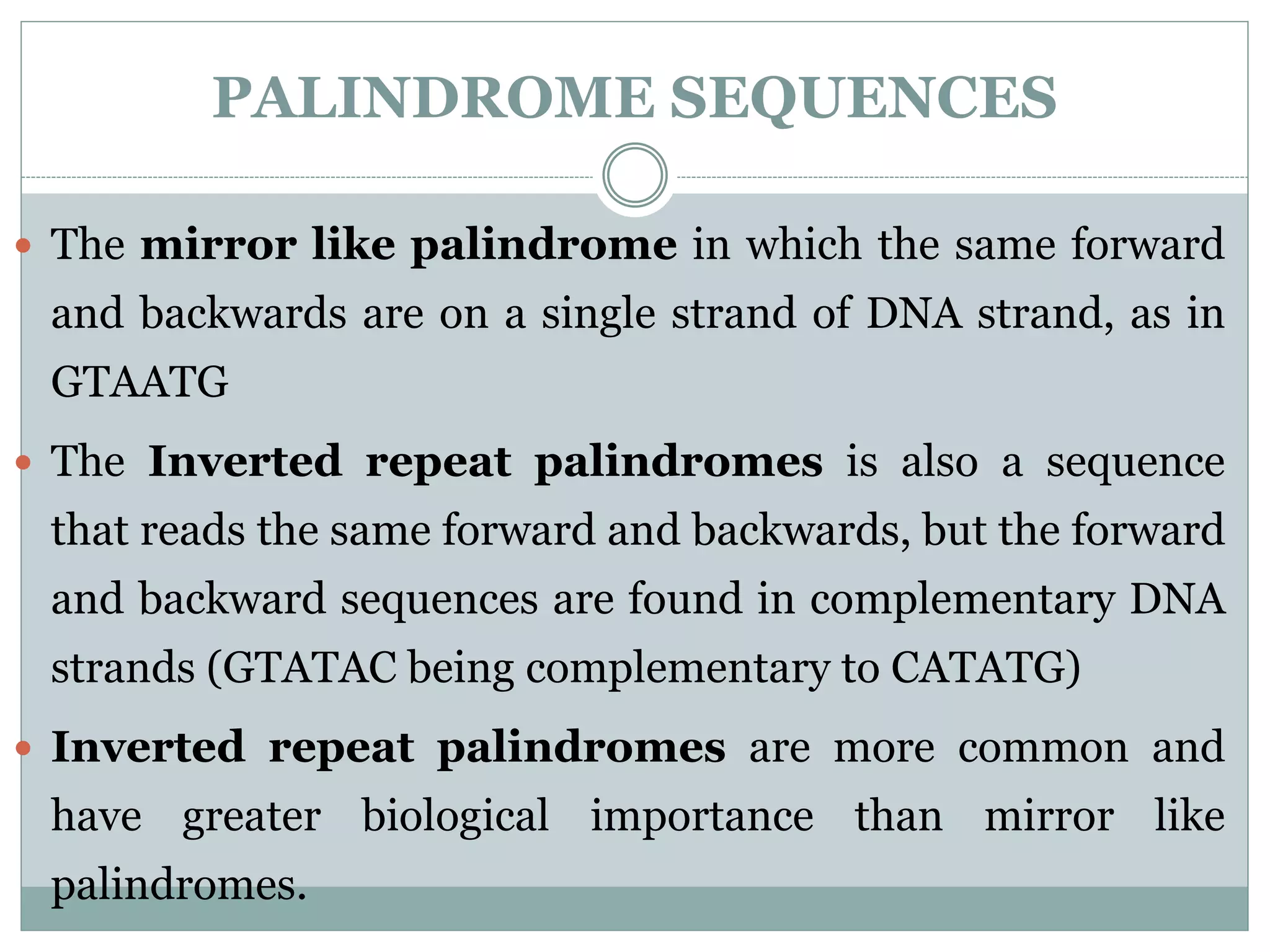
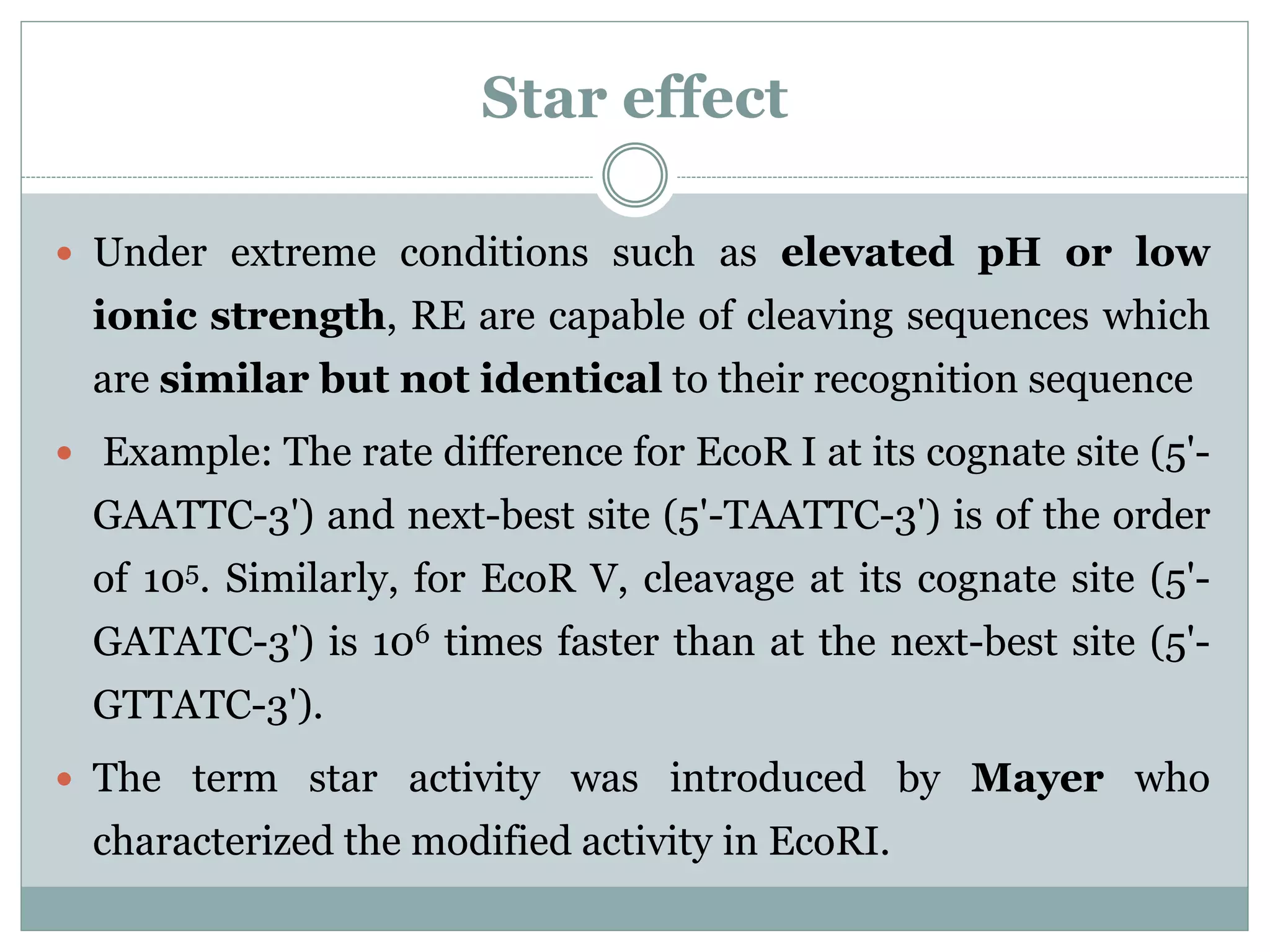
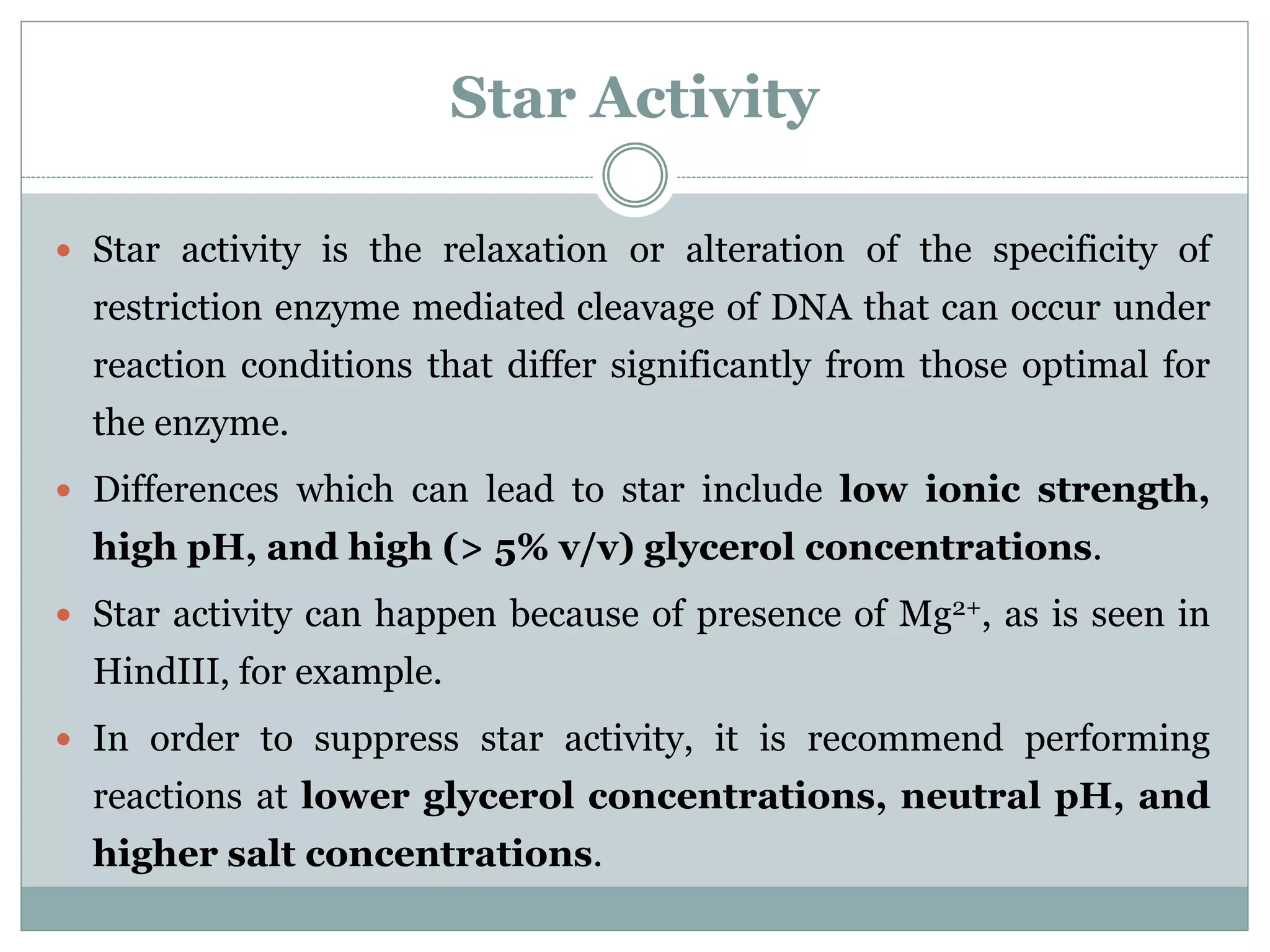
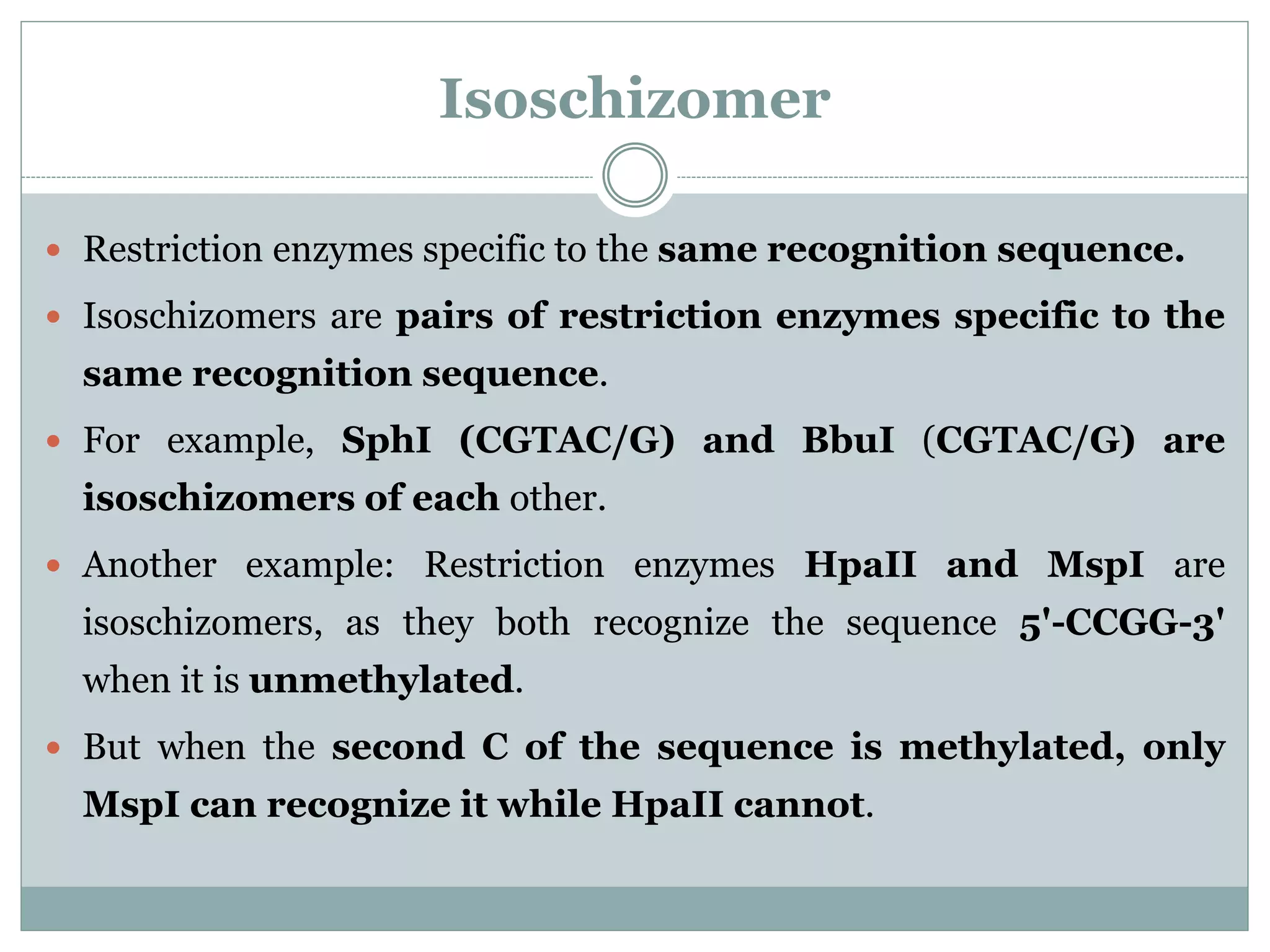
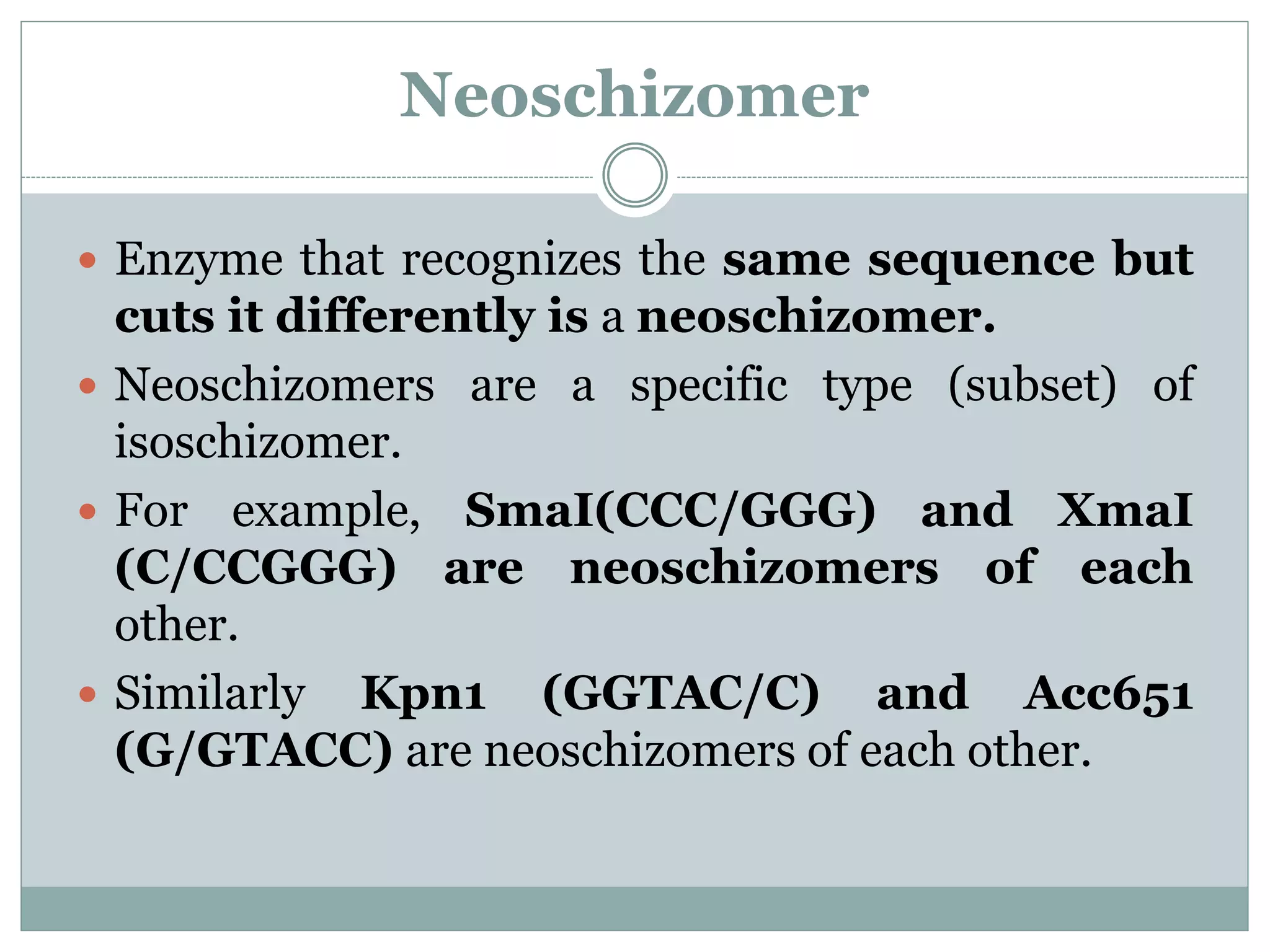
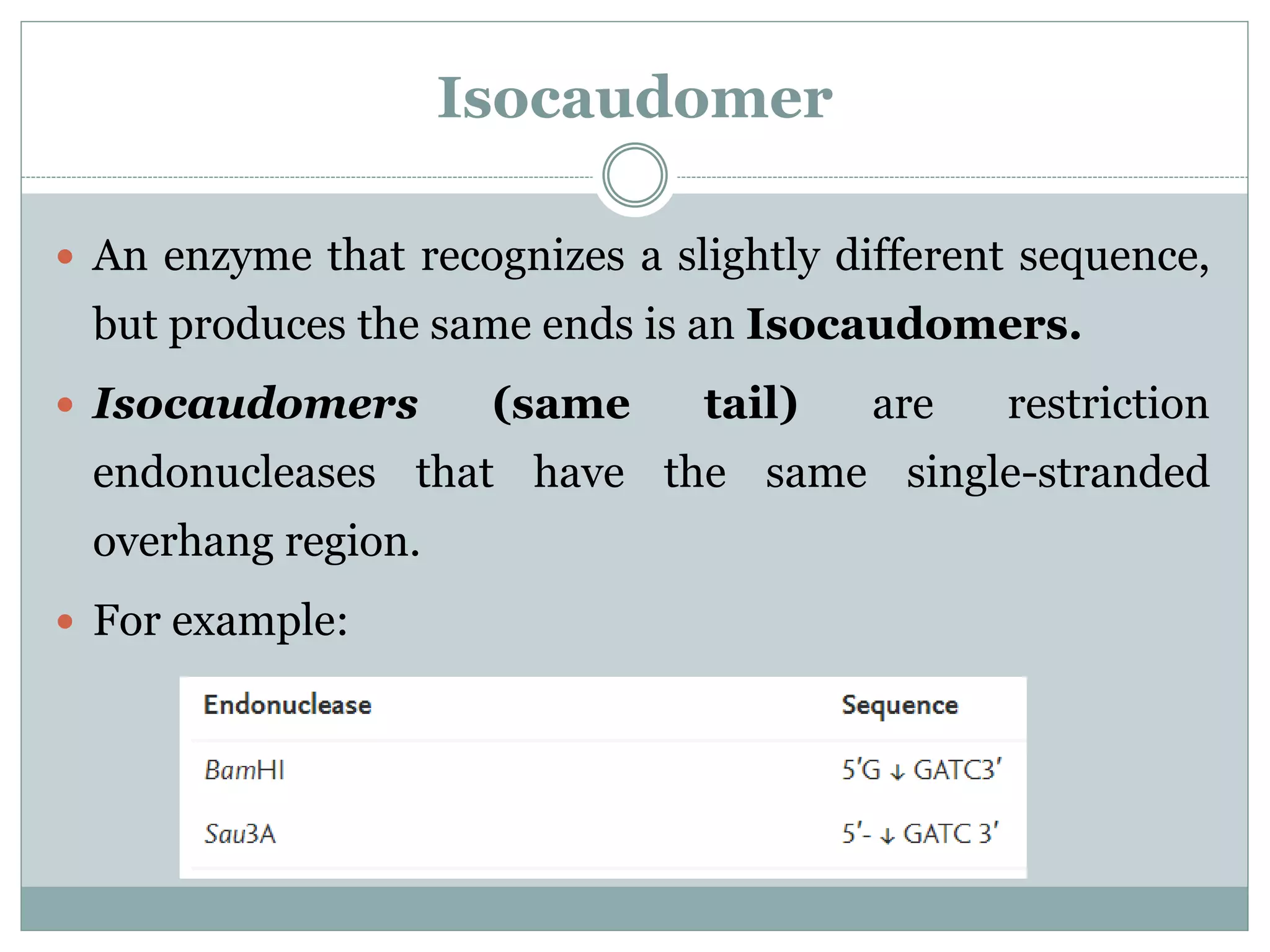
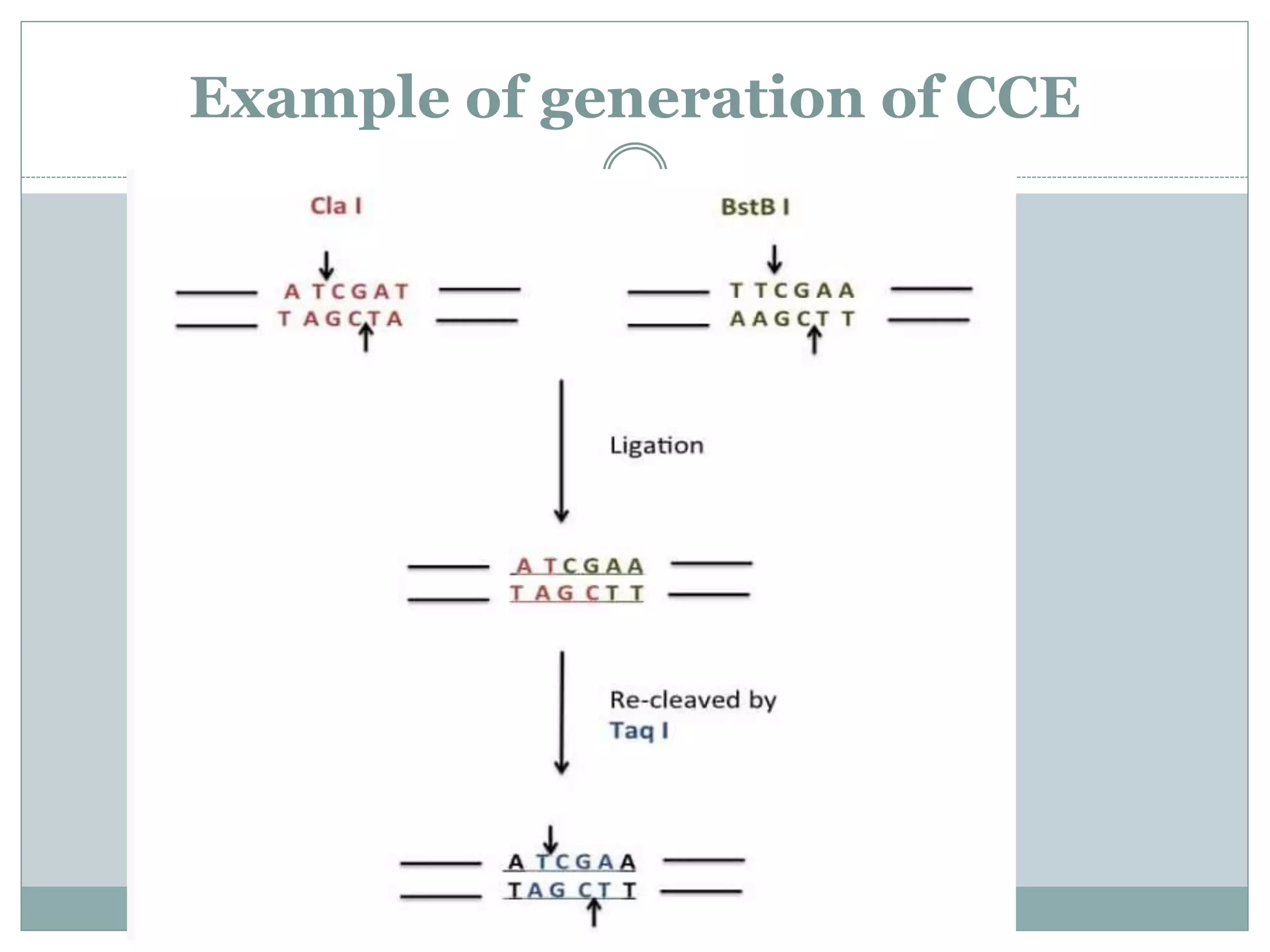
The restriction modification (RM) system is a bacterial defense mechanism against foreign DNA, utilizing restriction enzymes to cut DNA at specific sequences and modification enzymes to protect the host's own DNA by methylation. There are different types of restriction enzymes (Type I, II, III, and IV) with varying complexities, recognition sequences, and mechanisms of action, which are essential tools in genetic manipulation and DNA analysis. Additionally, concepts like star activity, isoschizomers, neoschizomers, and isocaudomers describe variations in enzyme specificity and cutting patterns, highlighting their versatile applications in biotechnology.