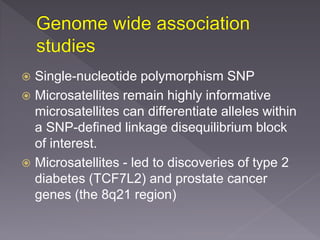
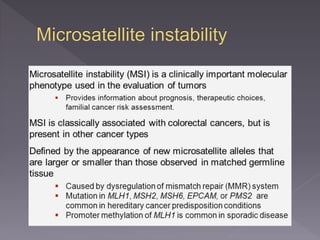
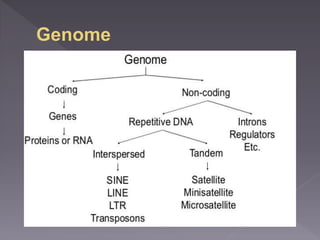
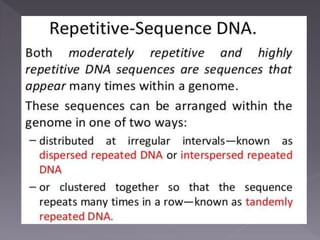
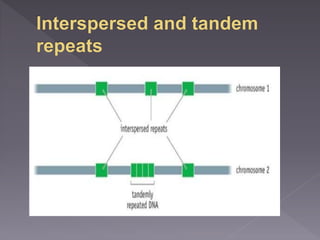
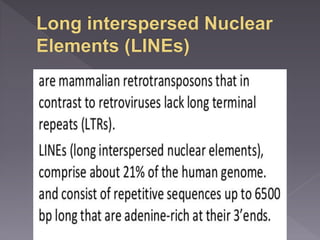
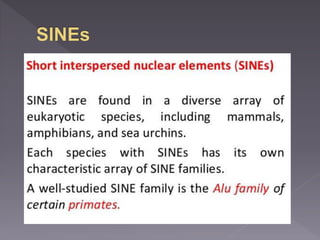
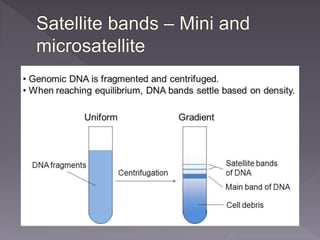
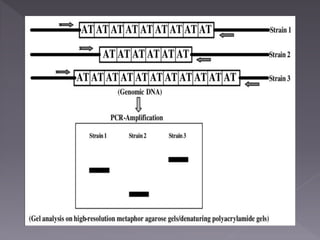
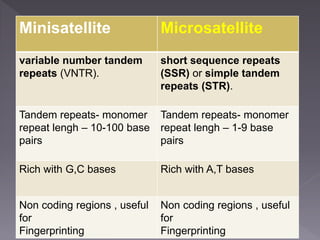
Microsatellites are short tandem repeats of DNA motifs between 1-9 base pairs found throughout genomes. They have high mutation rates and genetic diversity. Microsatellites are used for DNA fingerprinting, identifying individuals in forensics and paternity testing, and studying population genetics and genetic diseases like triplet expansion disorders.





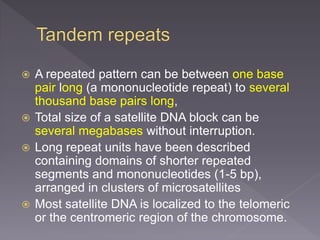


![ Microsatellites in non-coding regions allows
them to accumulate mutations unhindered
over the generations
So it gives rise to variability that can be
used for DNA fingerprinting and identification
purposes.
Some microsatellites are located in
regulatory regions of genes-
Mutations in such cases can lead to
phenotypic changes and diseases, notably
in triplet expansion diseases such as fragile
X syndrome and Huntington's disease.[9]](https://image.slidesharecdn.com/microsatellite-200608111600/85/Microsatellite-18-320.jpg)