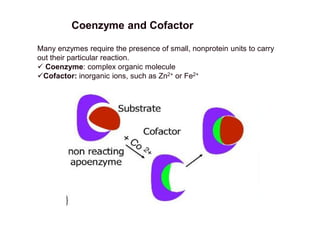
Enzymes are protein catalysts that speed up biochemical reactions without being consumed. They have specific three-dimensional structures with active sites that bind substrates and catalyze their conversion to products. There are six main enzyme classes defined by their catalyzed reaction types. Enzyme activity is affected by factors like substrate and enzyme concentration, pH, temperature, and inhibitors. Enzymes lower activation energy and use lock-and-key or induced-fit binding to catalyze reactions efficiently through transition states. Many require cofactors like vitamins and metal ions to function. Enzyme kinetics describe how rates change with conditions and inhibition mechanisms. Enzymes are essential to all living functions but some inhibitors like nerve gases are toxic.






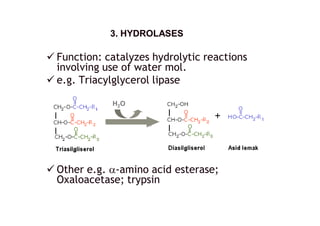
![ Function: catalyzes cleavage of C-C, C-O, C-
N and other bonds by other means than by
hydrolysis or oxidation.
e.g. Lysine decarboxylase
other e.g.: threonine aldolase [EC 4.1.2.5];
Other e.g. cystine lyase, pyruvate
decarboxylase
4. LYASES4. LYASES](https://image.slidesharecdn.com/enzymes-140413203918-phpapp01/85/Enzymes-8-320.jpg)
![ Function: catalyzes intramolecular transfer of
groups
e.g. Maleate isomerase
Other e.g. Inositol-3-phosphate synthase;
Maltose epimerase]
5. ISOMERASES5. ISOMERASES](https://image.slidesharecdn.com/enzymes-140413203918-phpapp01/85/Enzymes-9-320.jpg)