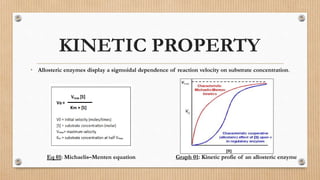
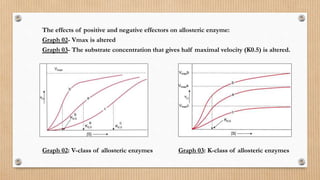
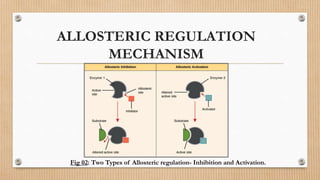
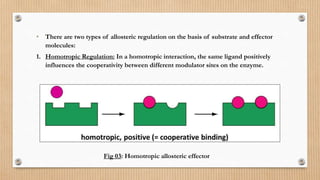
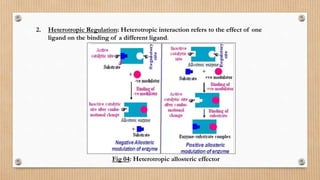
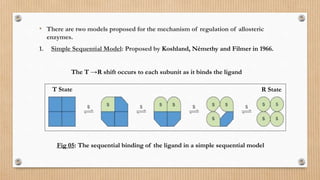
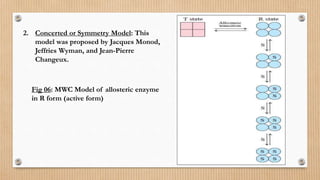
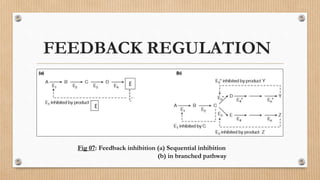
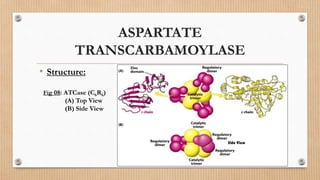
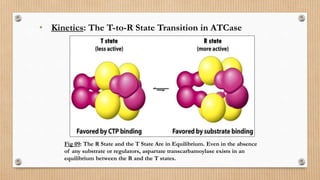
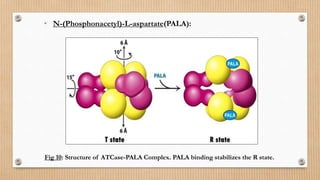
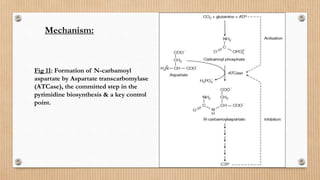
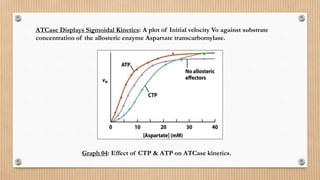
This document provides an overview of allosteric enzymes. It defines allosteric enzymes as enzymes whose activity is regulated by the binding of allosteric effectors at sites other than the active site. There are two types of allosteric effectors - positive effectors that increase enzyme activity and negative effectors that decrease it. Allosteric enzymes display cooperative binding and sigmoidal kinetics. They are classified as K-class or V-class depending on whether the effector changes the Km or Vmax value. Models like the Monod-Wyman-Changeux model and Koshland-Nemethy-Filmer model are described as proposed mechanisms for allosteric regulation. Aspartate transcarbamoylase