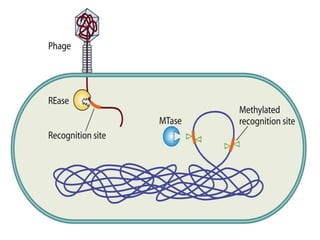
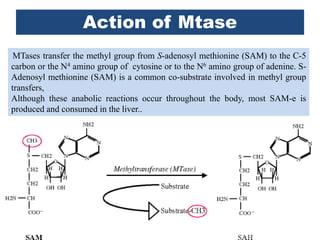
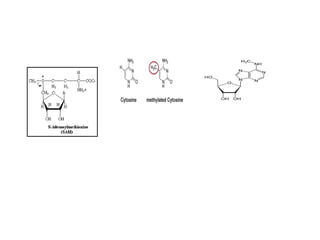
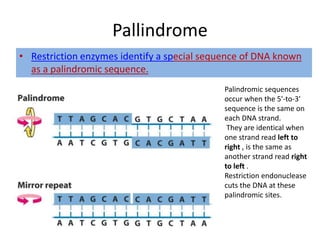
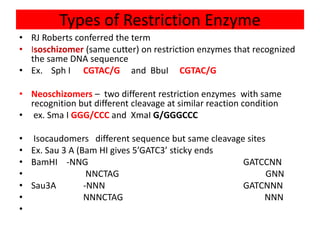
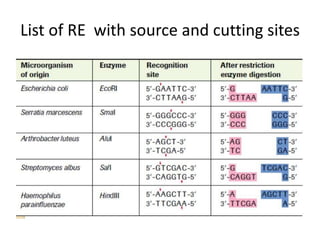
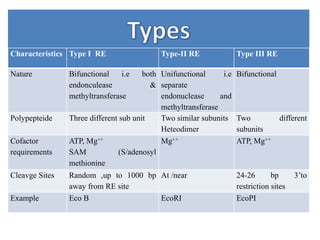
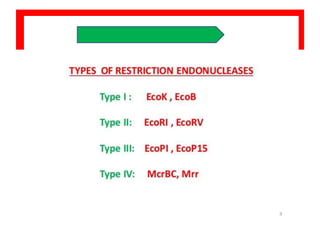
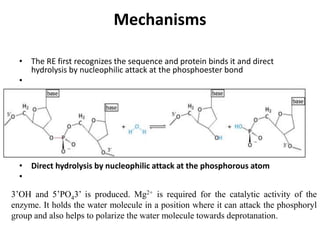
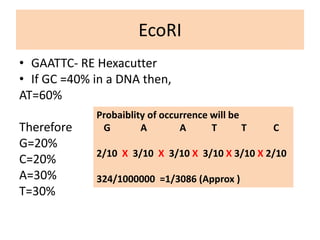
Restriction enzymes are proteins produced by bacteria that cleave DNA molecules at specific recognition sites. They play a role in bacterial host defense by cutting invading phage DNA but not the host's own DNA, which is protected by methylation. There are three types of restriction enzymes that differ in subunit structure and cofactor requirements. Type II restriction enzymes are the most commonly used molecular tools as they cut DNA into fragments at or near their recognition sites. The recognition sites are palindromic sequences that are cleaved by the restriction enzyme.