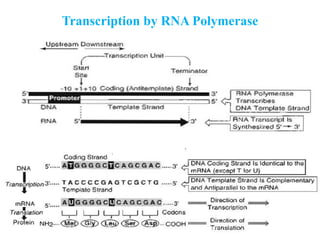
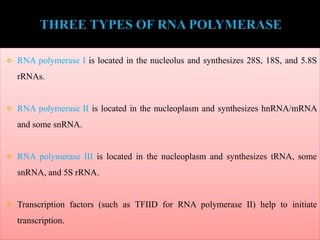
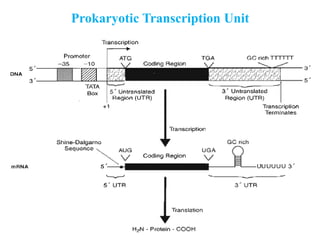
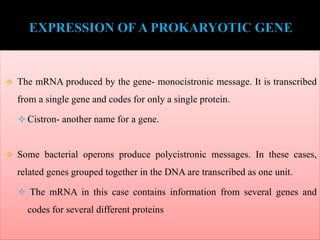
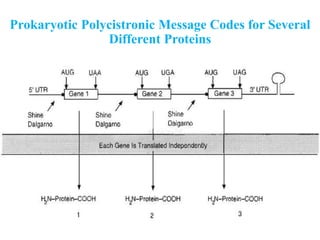
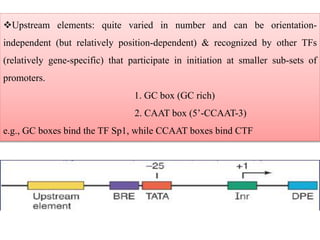
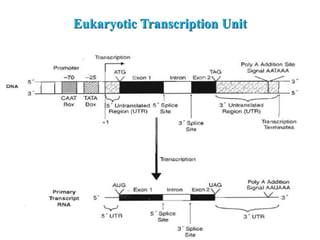
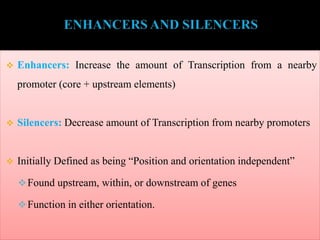
RNA polymerase (rnap) is essential for transcription, synthesizing RNA from DNA across all cells and is inhibited by specific compounds. It plays a crucial role in transcribing various types of RNA, with different polymerases (I, II, III) responsible for different RNA types in eukaryotes. Notably, research by Roger D. Kornberg has advanced understanding of rnap structure and function, including the discovery of a protein complex, 'mediator,' that aids in the transcription process.




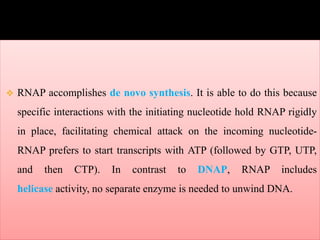
![Prokaryotic Eukaryotic
Bacterial Archaeal RNAP Ⅰ RNAP Ⅱ RNAP Ⅲ
Core Core (Pol Ⅰ ) (Pol Ⅱ) (Pol Ⅲ)
β‘ A’/A” RPC1 RPB1 RPC1
β B RPA2 RPB2 RPC5
α’ D RPC5 RPB3 RPC5
α“ L RPC9 RPB11 RPC6
ω K RPB6 RPB6 RPB6
[+6 other] [+9 others] [+7 others] [+11 others]](https://image.slidesharecdn.com/rnapolymerase-150507120506-lva1-app6891/85/Rna-polymerase-9-320.jpg)