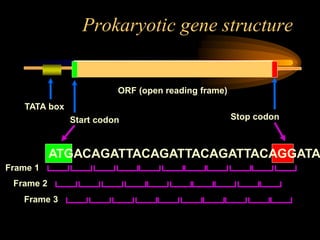
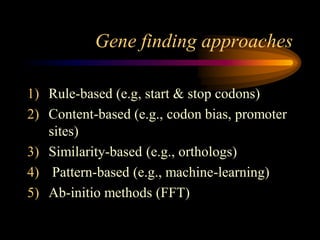
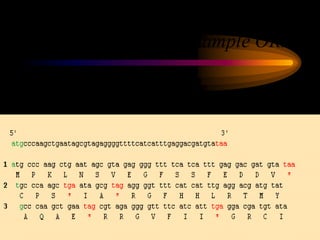
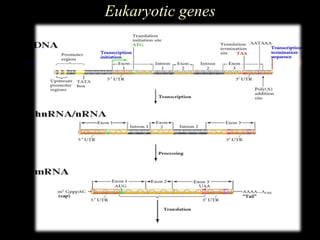
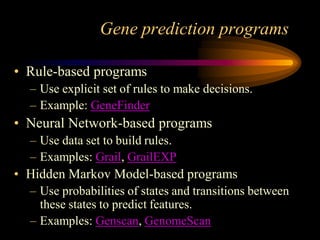
The document discusses gene prediction methods for prokaryotic and eukaryotic genes, highlighting the advantages and disadvantages of each approach. It details various gene finding strategies including rule-based, content-based, similarity-based, pattern-based, and ab-initio methods, as well as the use of machine learning techniques. Additionally, it reviews specific gene prediction programs and emphasizes the importance of combining multiple methods for improved accuracy.