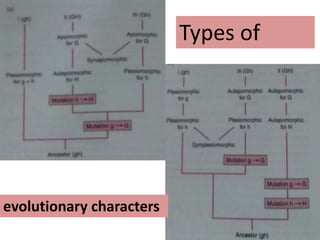
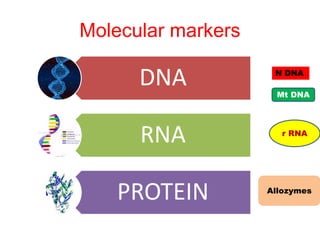
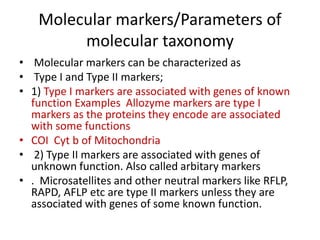
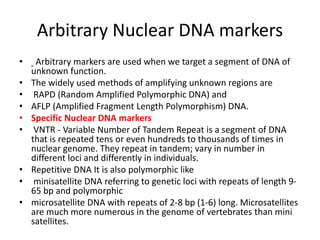
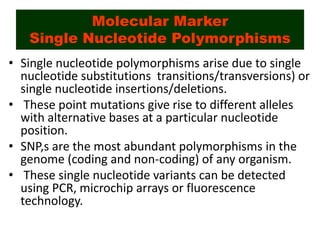
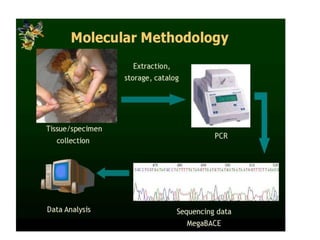
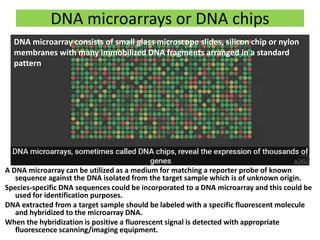
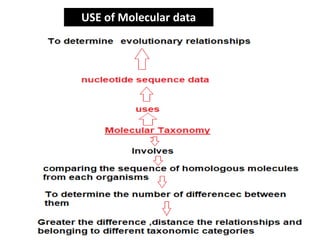
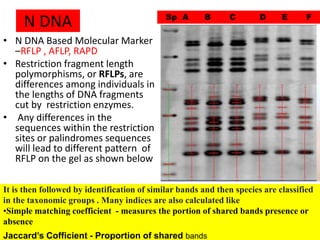
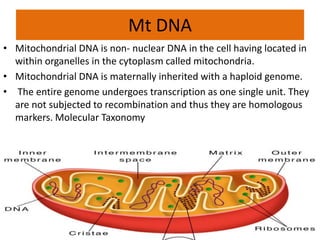
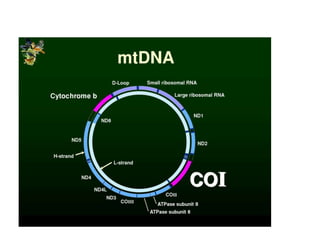
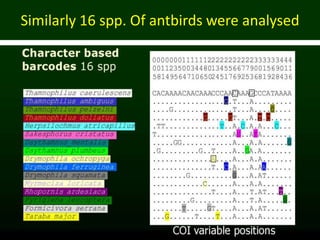
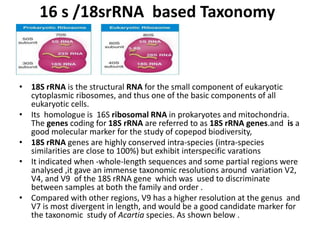
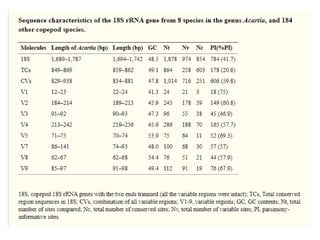
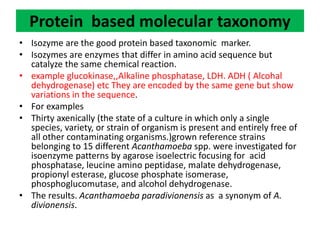
This document discusses molecular taxonomy and the use of molecular markers for classifying organisms. It describes how taxonomy has shifted from morphology-based to molecular-based as technology has advanced. Molecular markers like DNA, RNA, proteins, and allozymes can be used as they change at the microlevel during speciation. Common molecular markers discussed include mitochondrial DNA, rRNA, RFLPs, microsatellites, and isozymes. Techniques used include PCR, gel electrophoresis, and DNA microarrays. Examples are provided of various studies using molecular markers like COI, rRNA, and isozymes to classify species of bacteria, birds, and protozoa. Molecular taxonomy is concluded to be more accurate than morphology-based taxonomy as