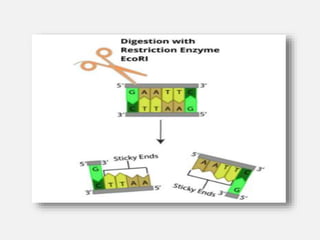
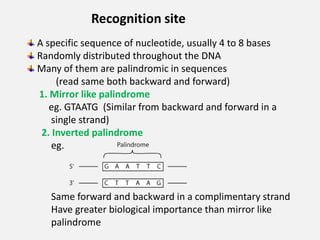
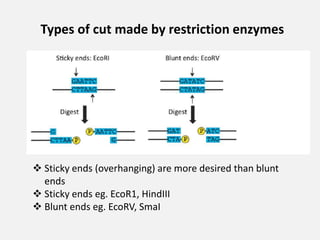
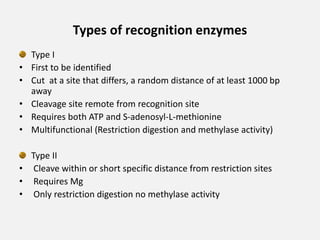
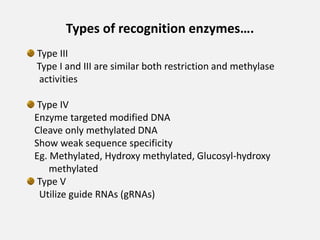
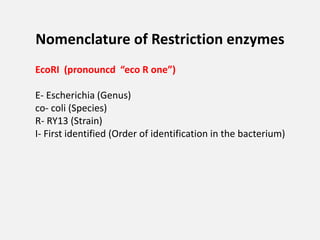
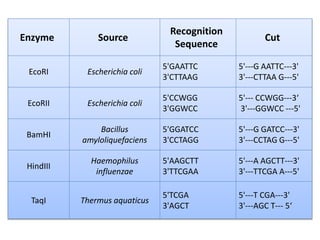
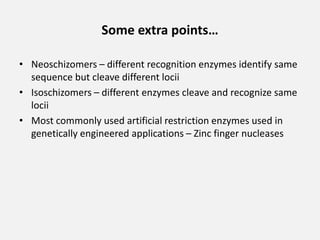
Restriction enzymes are proteins isolated from bacteria that cleave DNA into fragments at specific recognition sites. They recognize short palindromic nucleotide sequences, usually 4-8 bases long, and cut through both strands of the DNA double helix. There are four main types of restriction enzymes that differ in their recognition sites and cleavage activities. Restriction enzymes are indispensable tools in recombinant DNA technology, as they allow DNA fragments to be cut and recombined in new configurations.