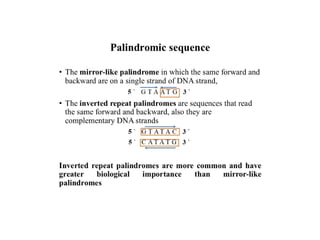
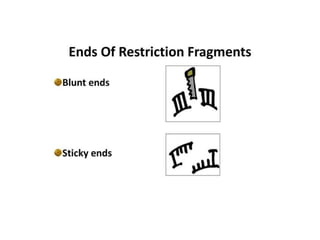
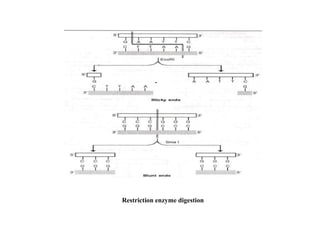
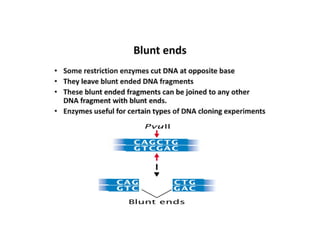
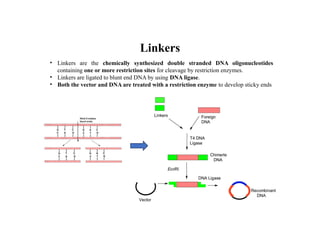
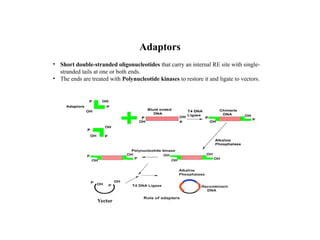
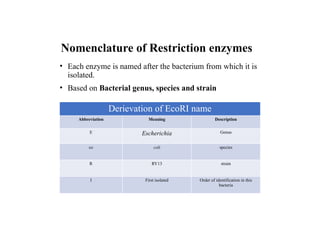
Restriction endonucleases are categorized into various types based on their composition, cofactor requirements, target sequence nature, and cleavage positions. Sticky ends, linkers, and adaptors are important tools for creating recombinant DNA, while different enzyme types, such as Type I, II, and III, exhibit distinct characteristics in recognition and cleavage of DNA. The document also details the nomenclature of restriction enzymes based on their bacterial origins and provides examples of each type.