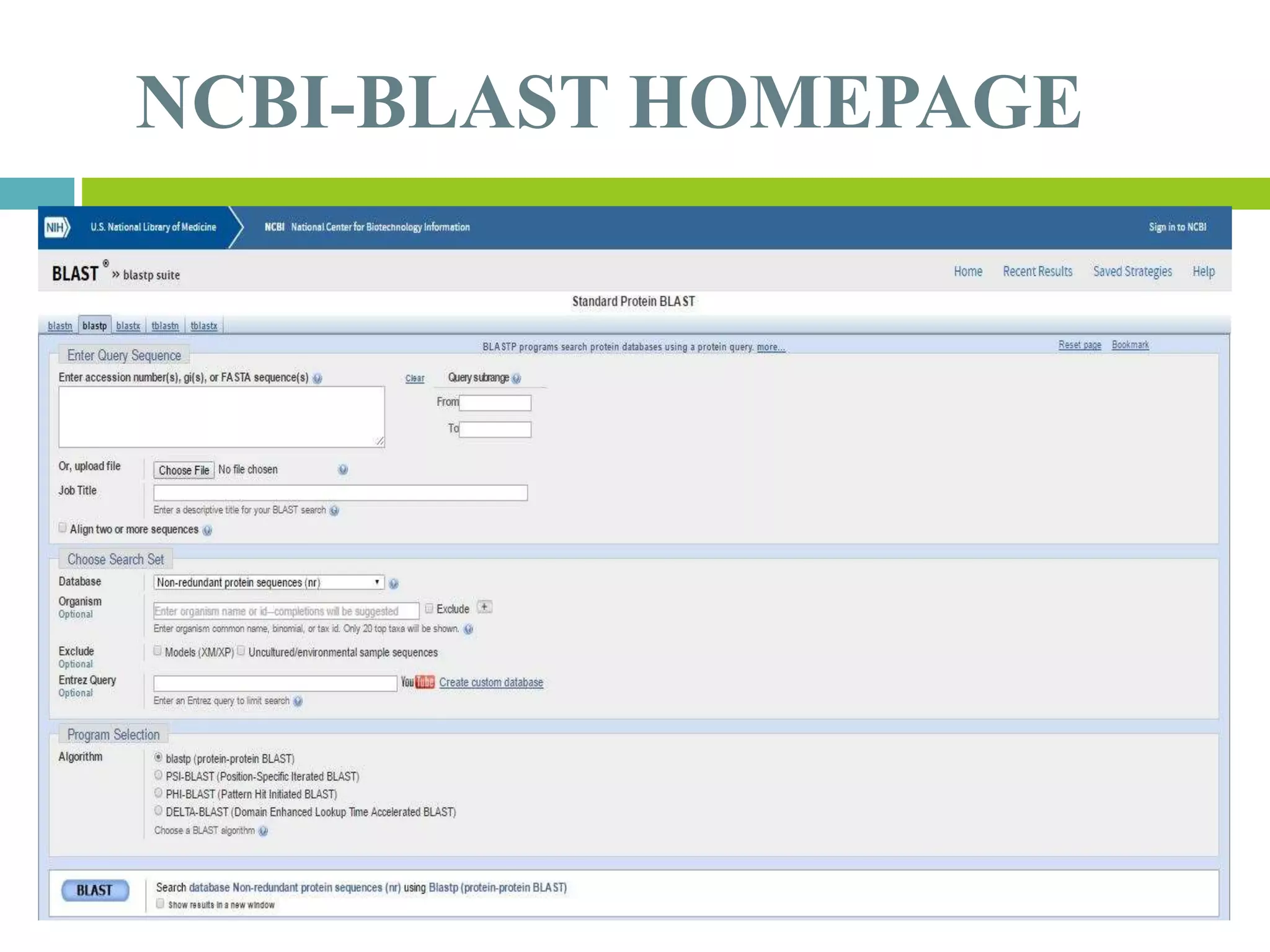
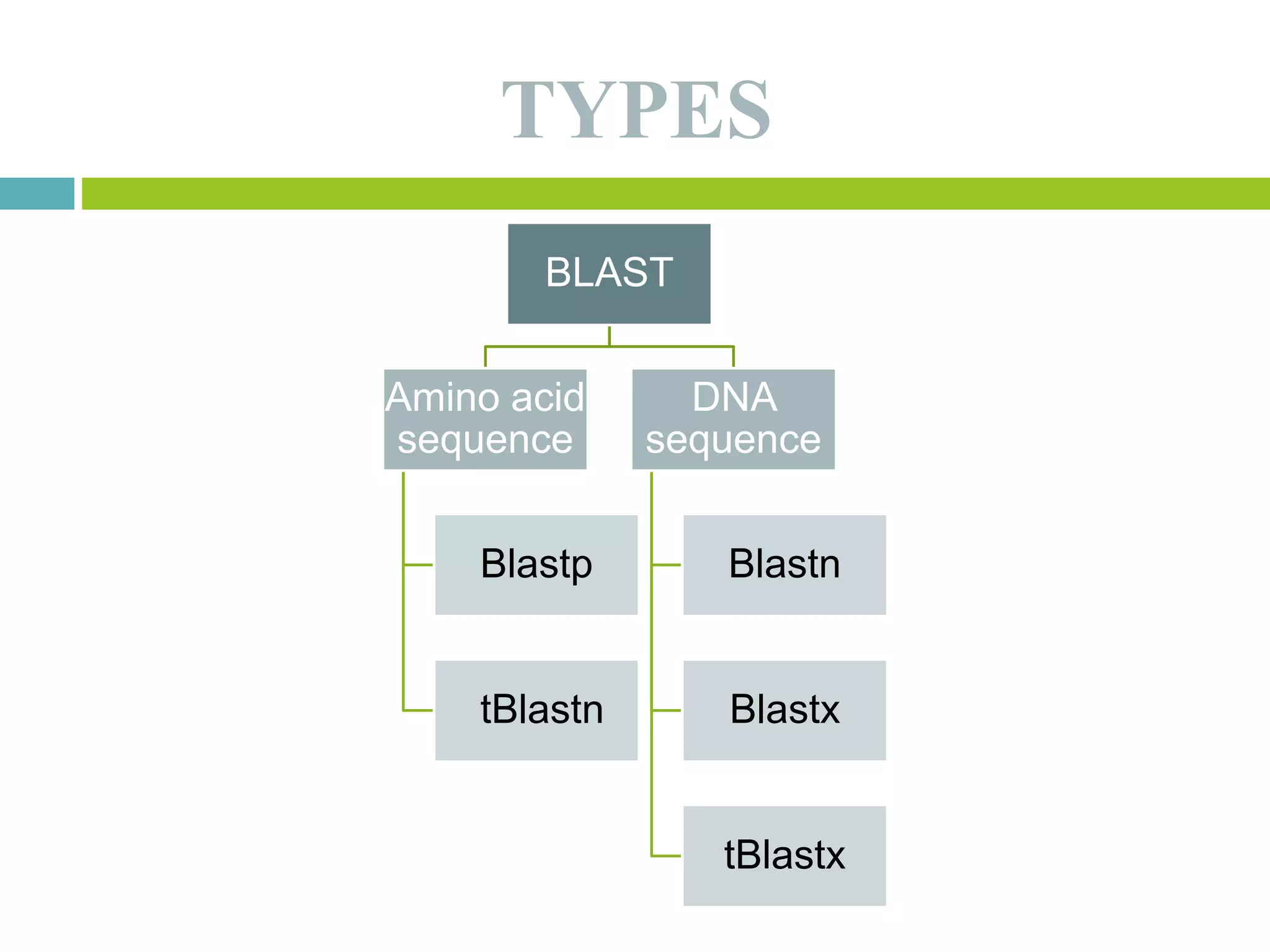
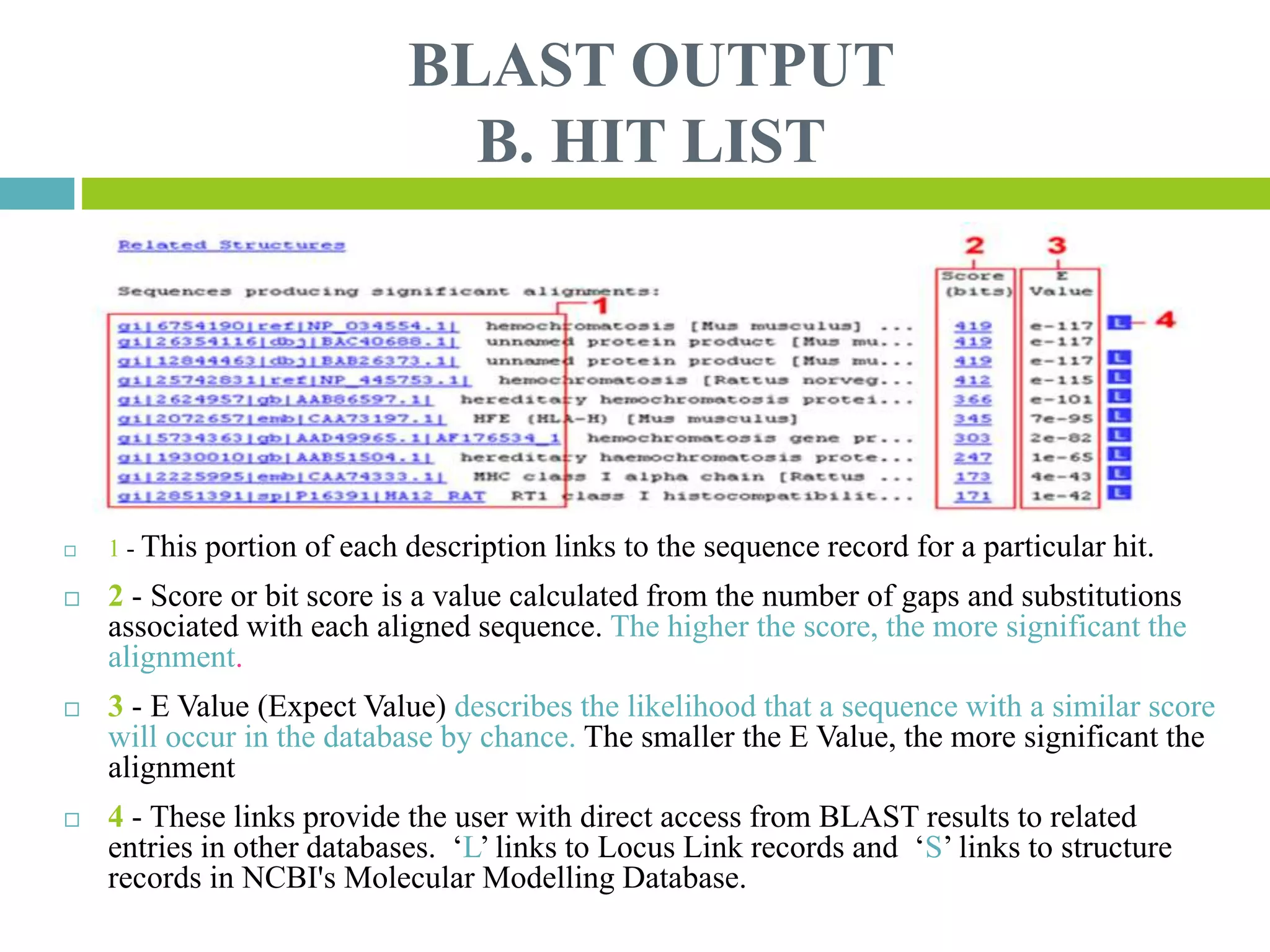
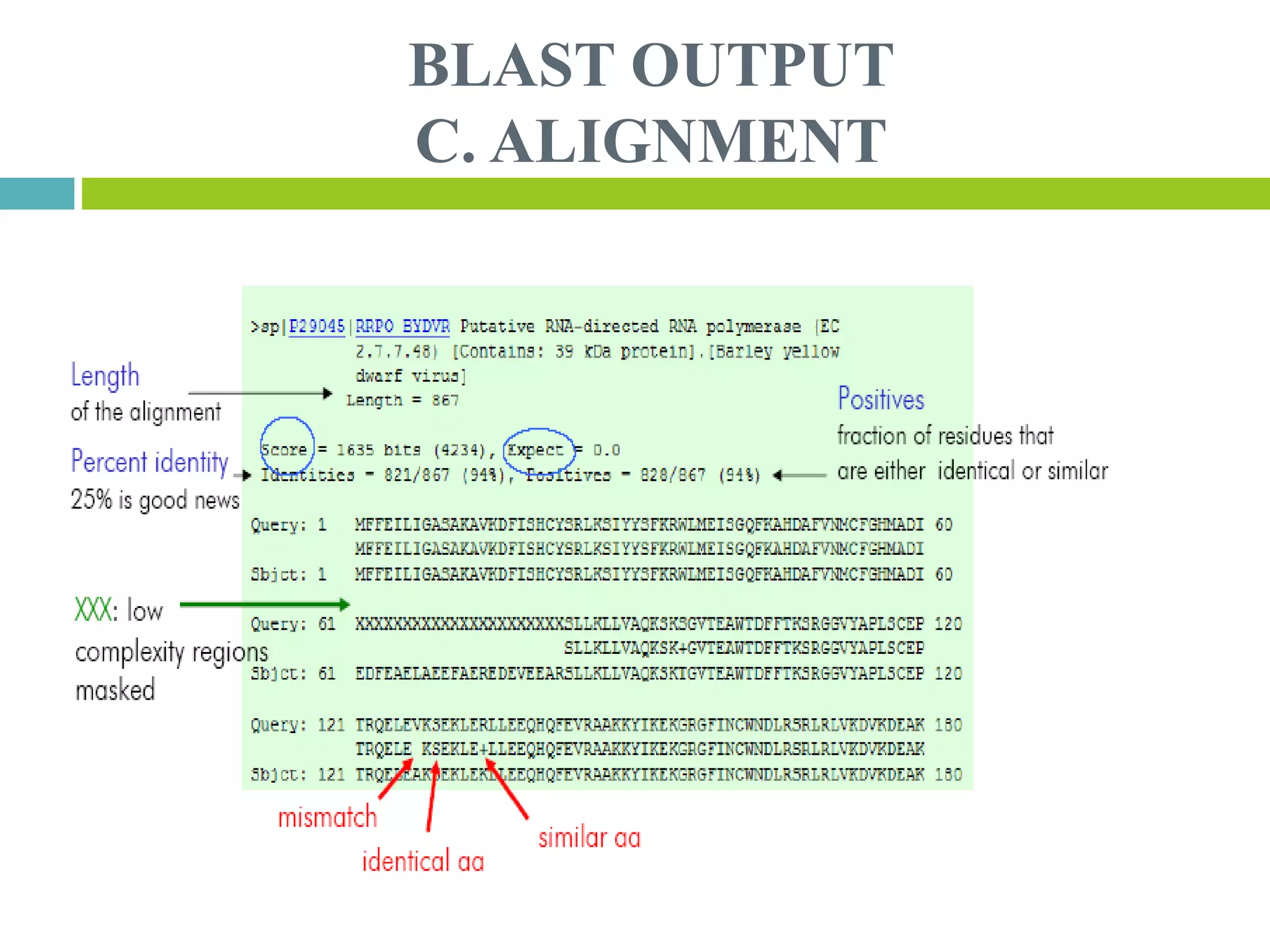
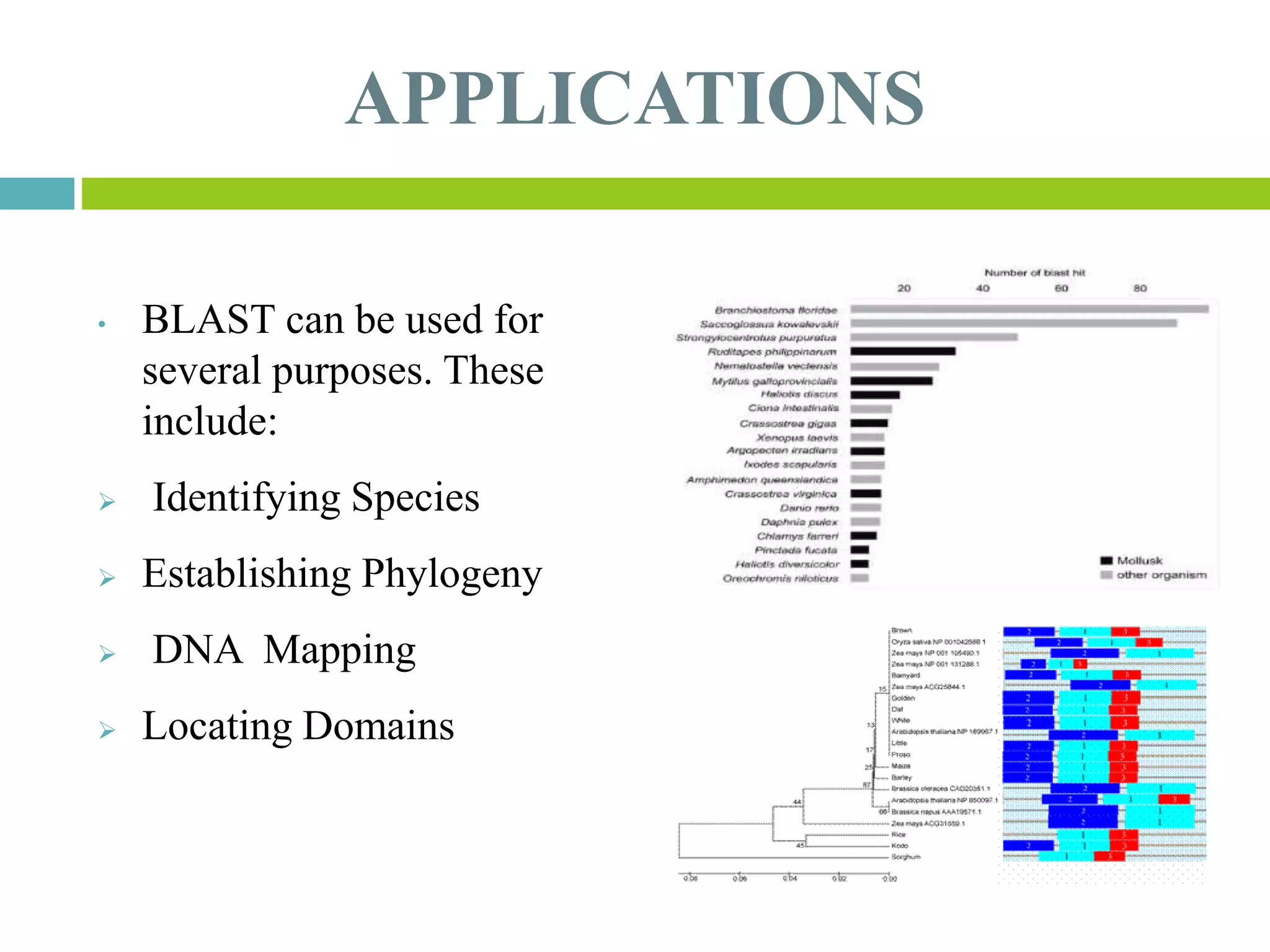
This document discusses the Basic Local Alignment Search Tool (BLAST), which allows users to compare a query DNA or protein sequence against sequence databases to find regions of local similarity. BLAST breaks the query into short words that are then searched for in database sequences. When words are found in common, BLAST extends the alignment in both directions to find higher-scoring matches. BLAST outputs include a graphical display of alignments, a hit list ranking matches by similarity score, and detailed alignments. BLAST has many applications, such as identifying species, establishing evolutionary relationships, DNA mapping, and locating protein domains.