Embed presentation
Download to read offline

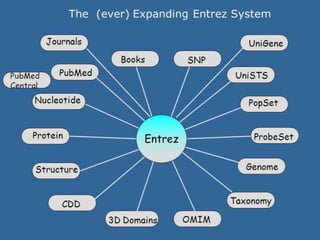
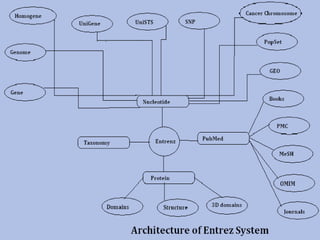
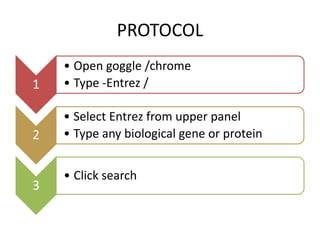


Entrez is a search engine developed by the National Center for Biotechnology Information (NCB) that allows users to search and retrieve data from over 20 integrated biological databases, including sequences, gene records, citations, and abstracts. It provides a single interface to access all linked information on genes and proteins. Users can perform text searches using Boolean operators and view results in various formats like FASTA and XML.








