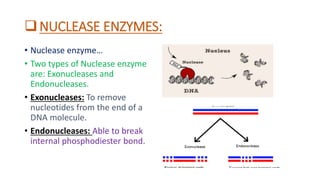
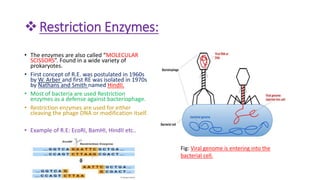
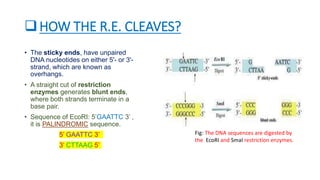
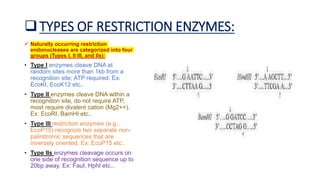
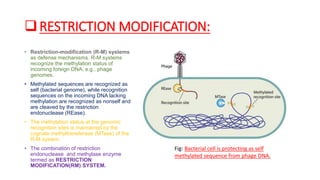
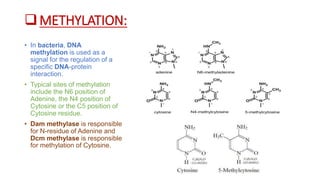
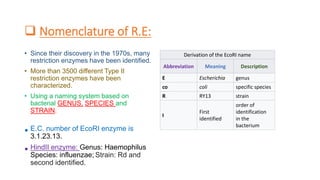
Restriction enzymes, also known as molecular scissors, are nuclease enzymes found in prokaryotes that cut DNA at specific recognition sites. They were first discovered in the 1960s and are used by bacteria as a defense against bacteriophages by cleaving or modifying viral DNA. There are four main types of restriction enzymes that differ in their recognition sites, cleavage patterns, and enzymatic requirements. Restriction enzymes are widely used in biotechnology and molecular biology applications such as DNA cloning, gene analysis, and production of therapeutic proteins.