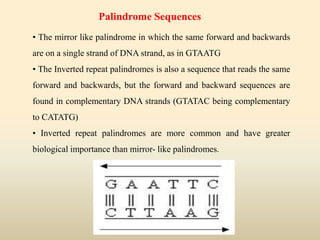
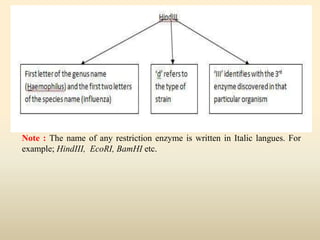
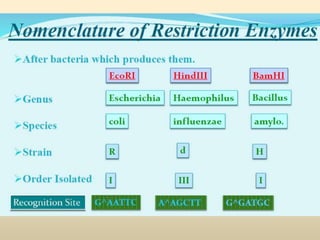
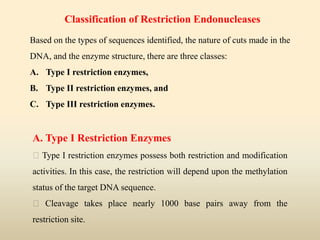
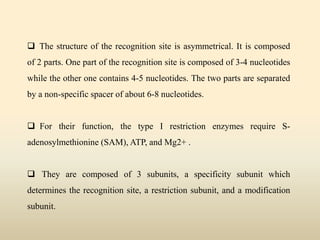
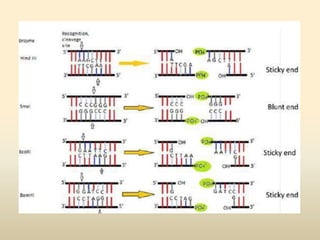
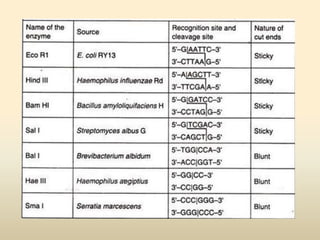
This document provides a comprehensive overview of restriction enzymes, including their definitions, types (I, II, and III), classification, and biological roles. It details the discovery history, structure, function, nucleotide cleavage patterns, and applications such as gene cloning and SNP analysis. Key terms and concepts such as palindromic sequences, isoschizomers, and star activity are also explained.