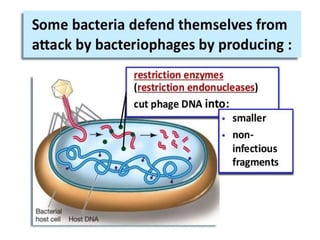
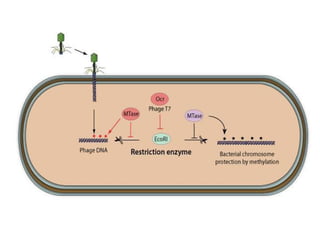
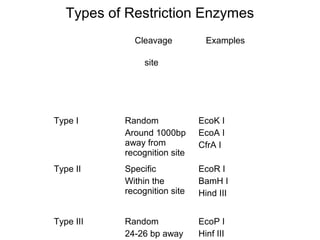
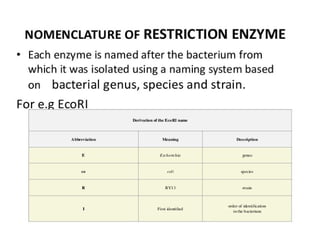
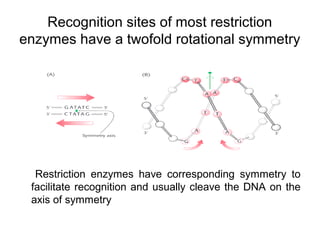
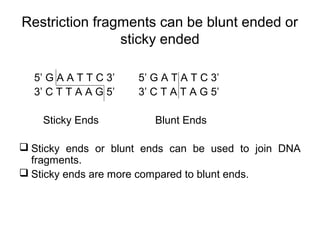
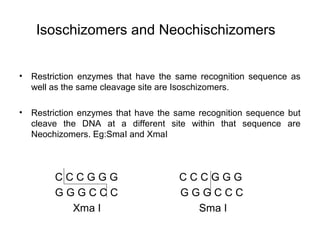
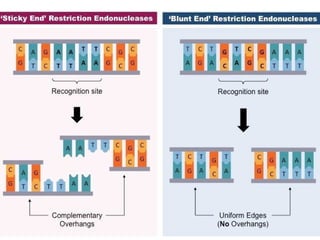
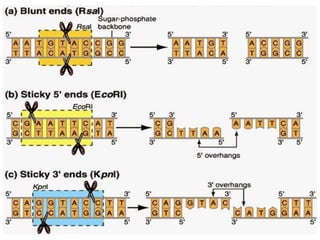
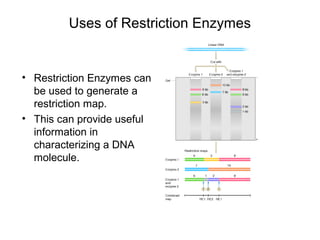
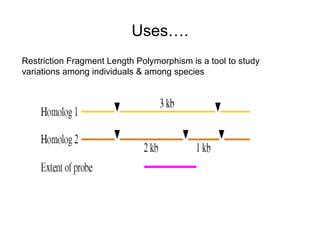
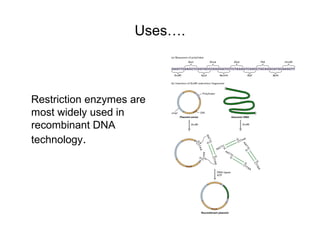
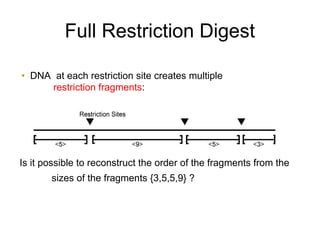
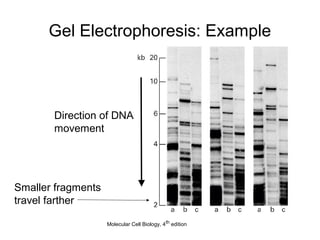
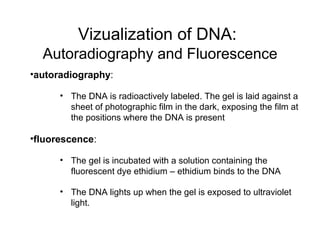
Restriction enzymes are molecular scissors that cut DNA at specific recognition sequences. They are found in bacteria and archaea and serve to defend the host genome from foreign DNA like viruses. There are four main types of restriction enzymes that differ in composition, cofactor requirements, target sequence, and cleavage site. Restriction enzymes are important tools in biotechnology for manipulating DNA through cutting and joining of fragments. They allow for processes like DNA cloning, mapping, and sequencing.