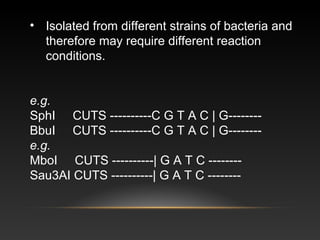
This document discusses isoschizomers, neoschizomers, and isocaudomers - types of restriction enzymes that recognize the same or similar DNA sequences but may cut the DNA differently. It provides examples like SphI and BbuI which are isoschizomers that recognize the same sequence (CGTAC/G) but were isolated from different bacteria. It also discusses the uses of restriction enzymes in recombinant DNA technology and their biological role in bacteria to modify or destroy incoming DNA.