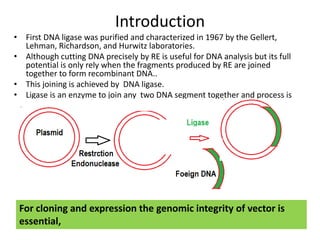
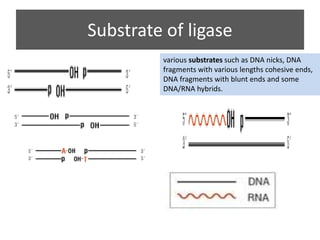
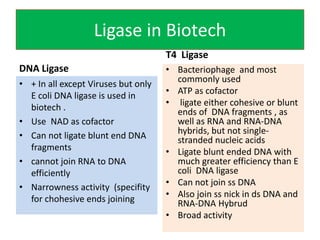
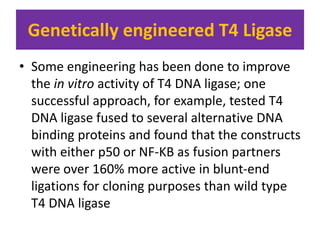
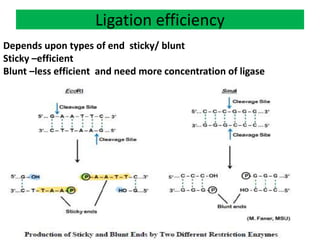
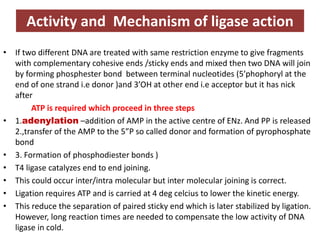
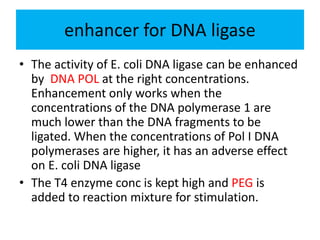
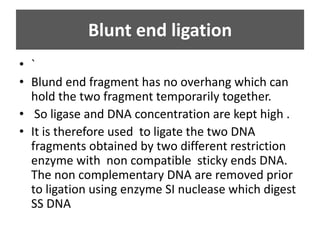
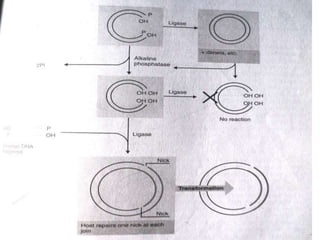
DNA ligase is an essential enzyme that joins together DNA fragments. The first DNA ligase was purified in 1967. It plays a key role in processes like DNA replication, repair, and recombinant DNA techniques. There are different types of ligases from sources like E. coli and T4 bacteriophage. Ligases can join DNA fragments with complementary sticky ends or blunt ends, and are used to integrate DNA into vectors for cloning. T4 ligase is most commonly used as it acts on a broad range of substrates.