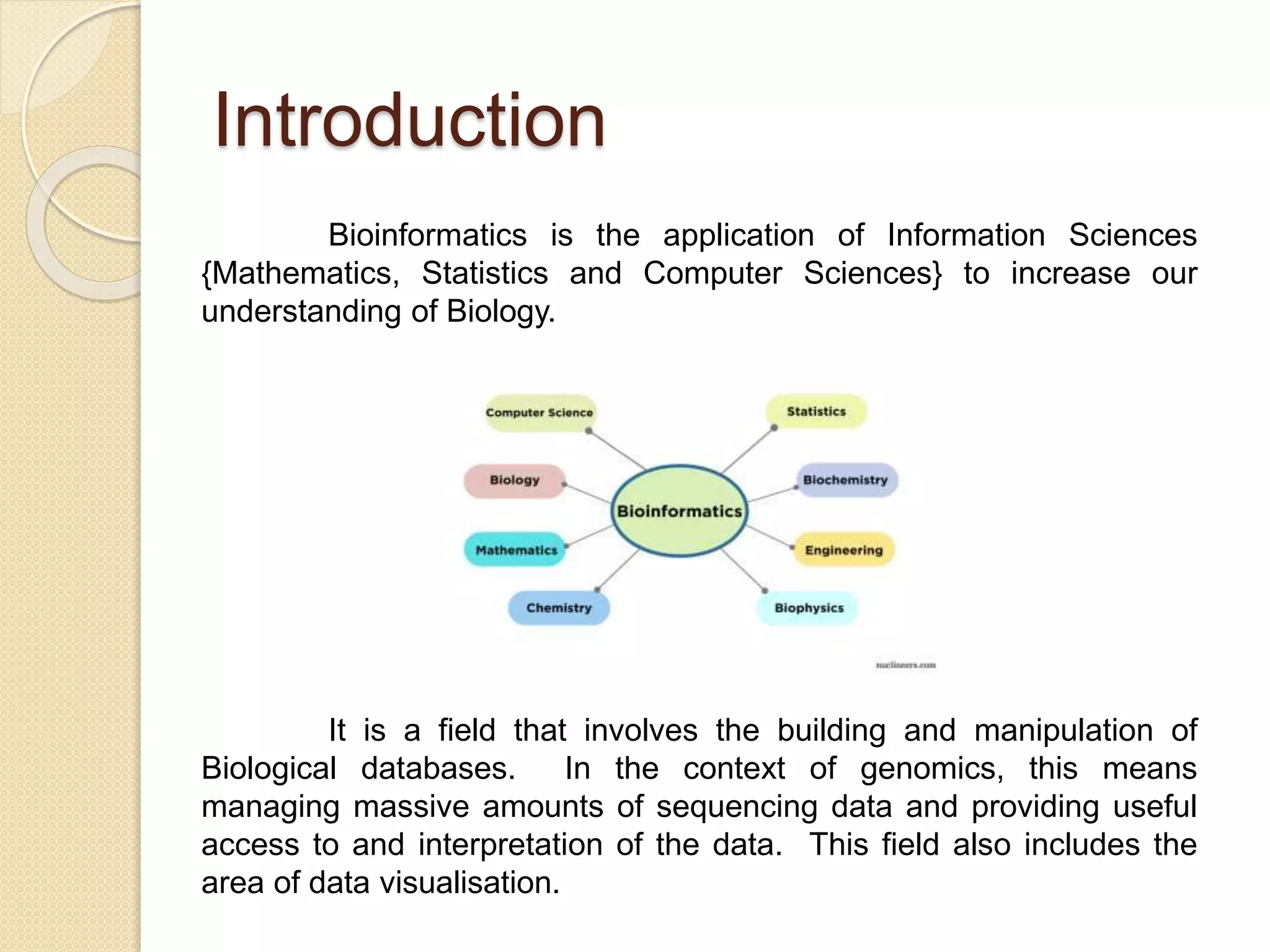
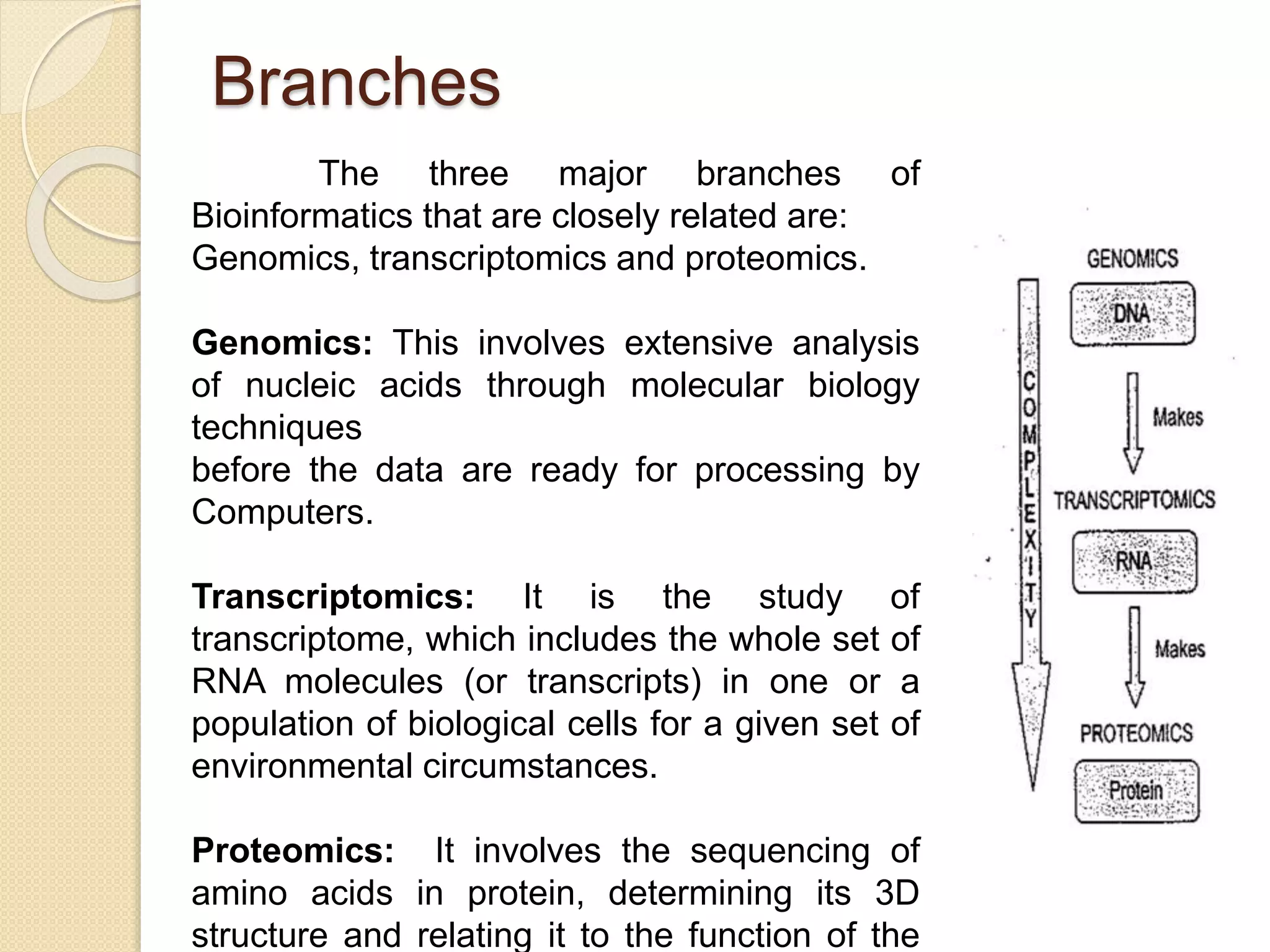
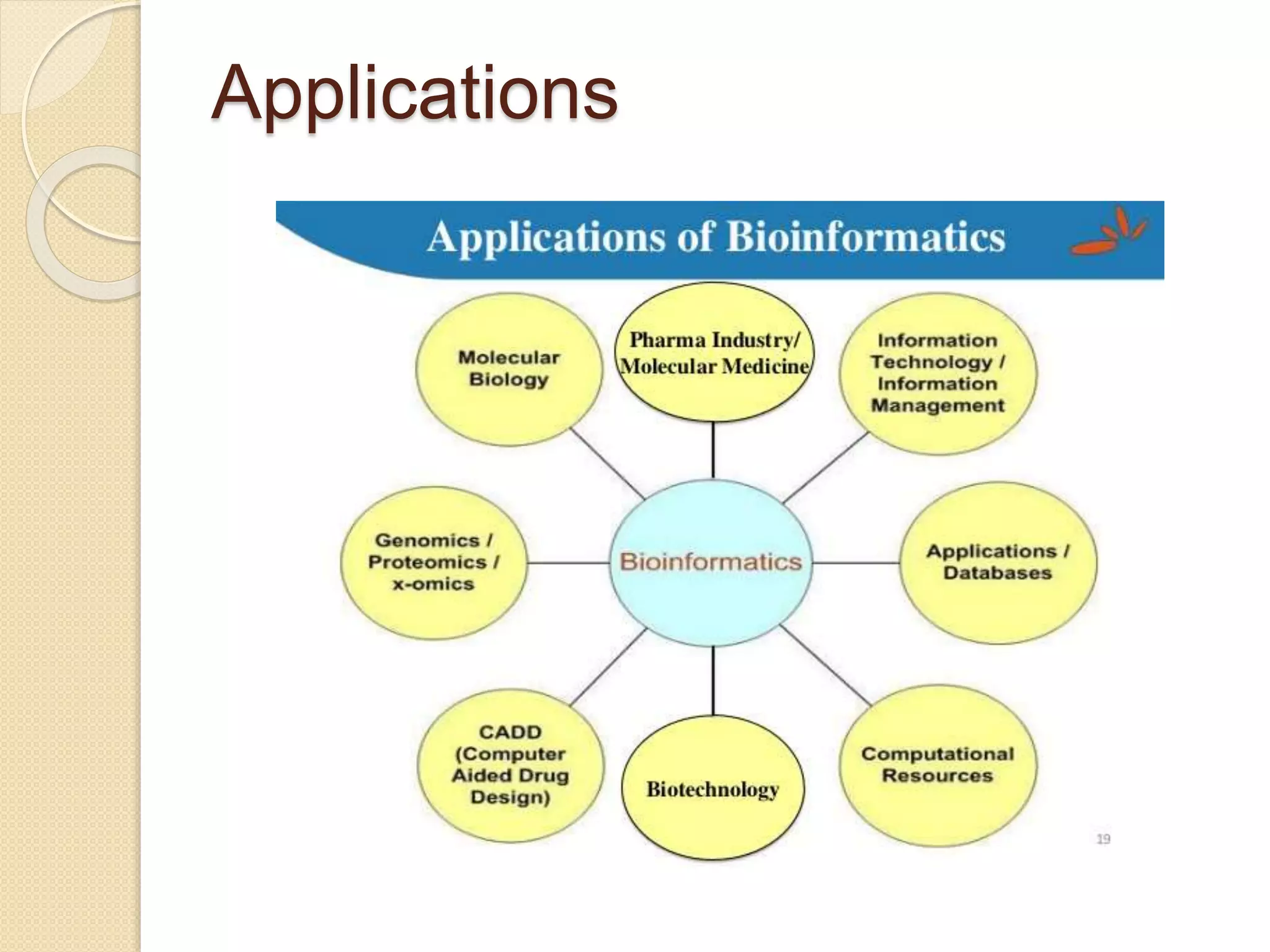
The document discusses the importance and scope of bioinformatics, highlighting its role in managing biological data in the post-genomic era. It outlines the history of the field, beginning from the early studies in the 1940s to the establishment of major databases in the 1980s. Applications of bioinformatics are focused on genomics, transcriptomics, and proteomics, emphasizing its significance in biomedical research.