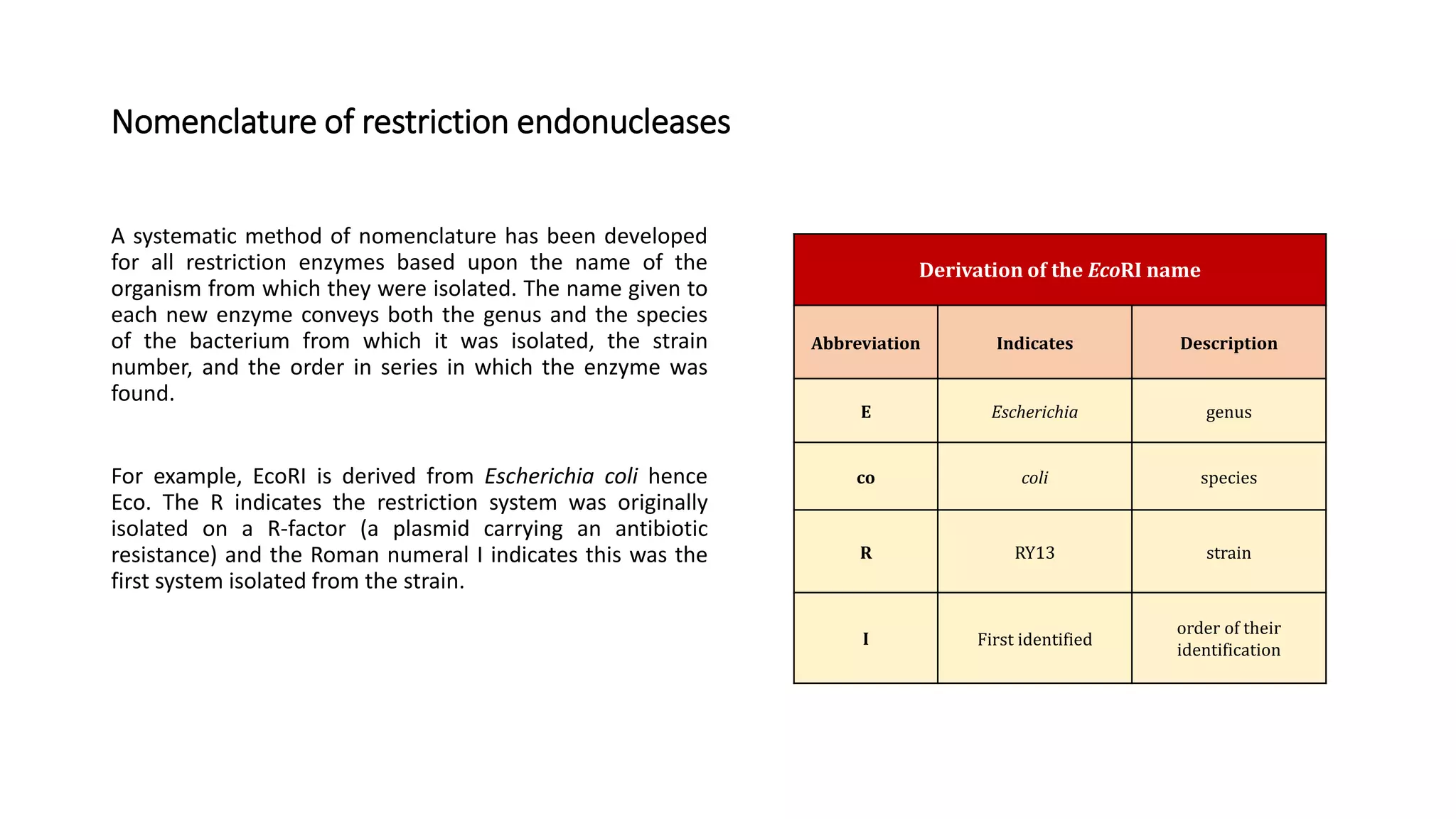
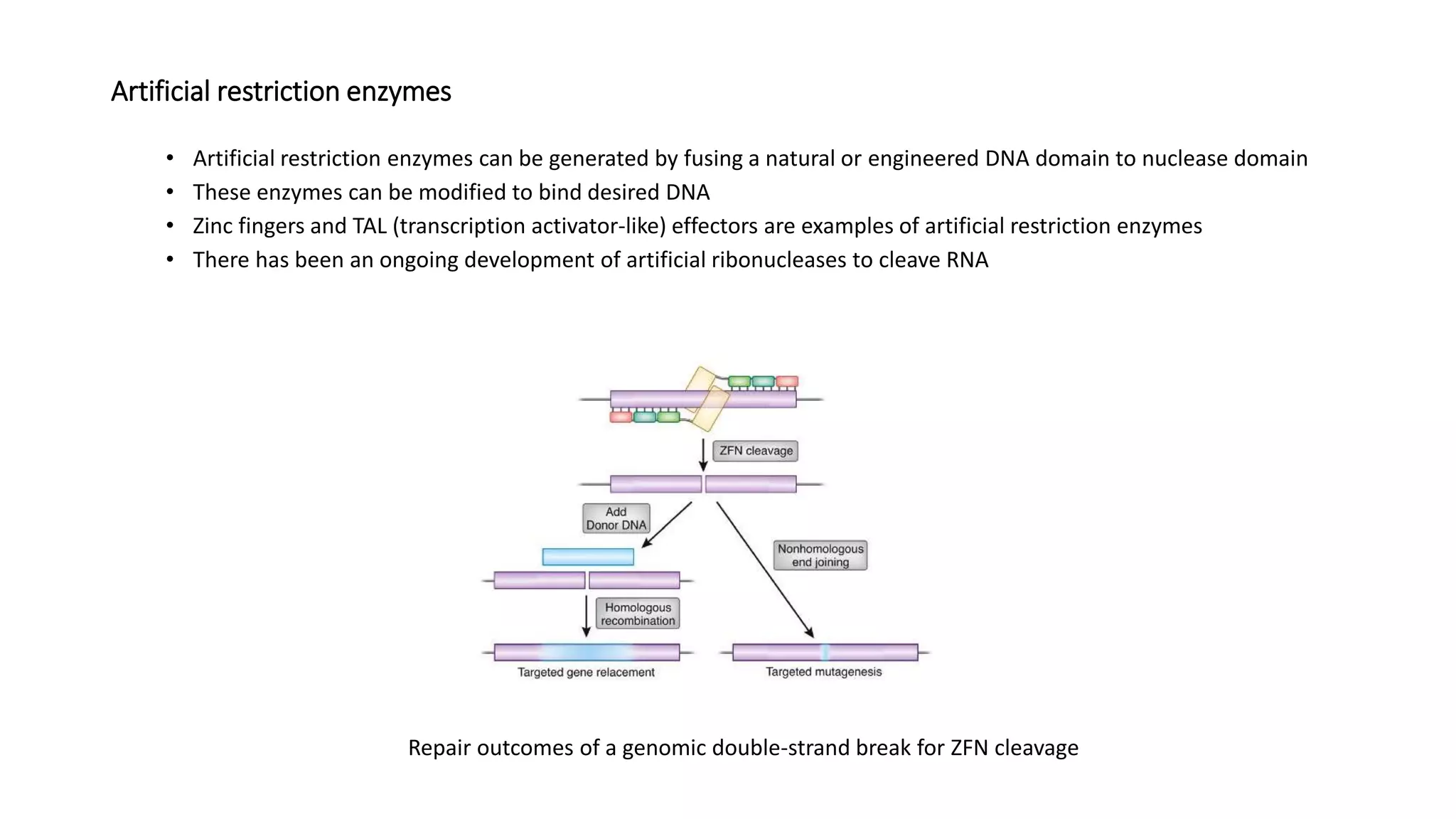
Restriction endonucleases are essential enzymes in molecular biology that modify nucleic acids, serving as a foundation for recombinant DNA technologies and biotechnology. These enzymes are categorized into different types based on their structure and mechanism, with type II enzymes being the most common for cutting DNA at specific sequences, facilitating cloning and genetic engineering. Their precise action allows for applications ranging from DNA mapping to in vivo gene editing and synthetic biology.