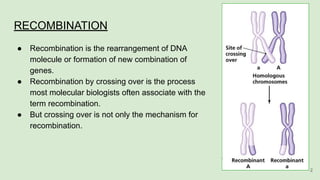
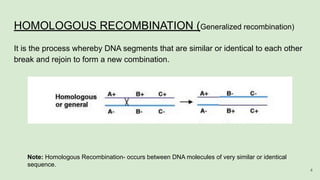
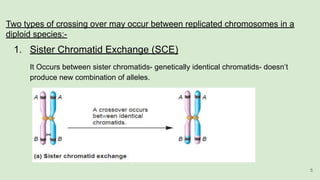
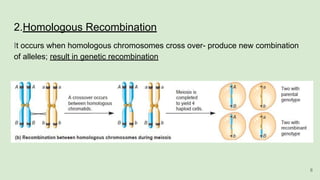
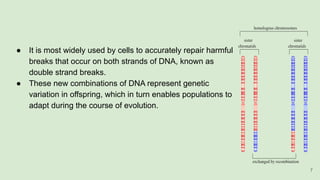
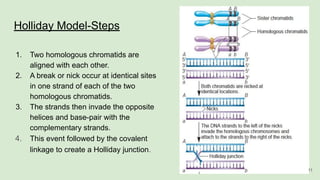
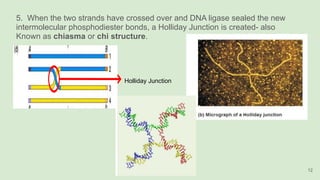
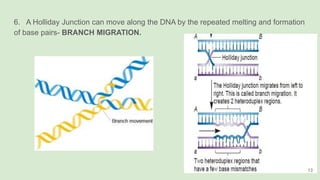
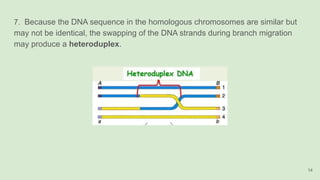
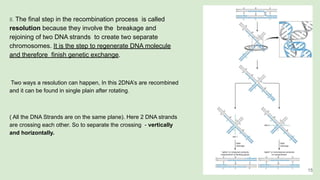
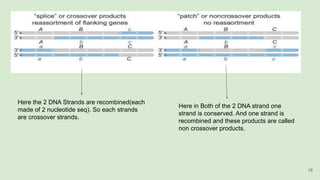
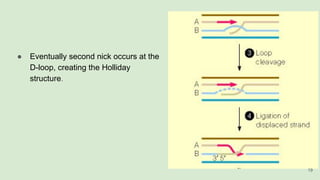
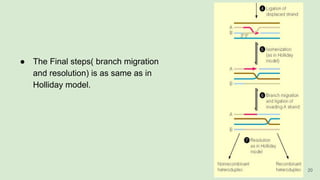
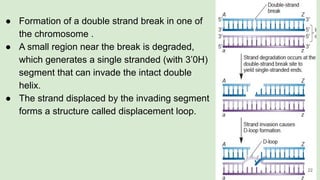
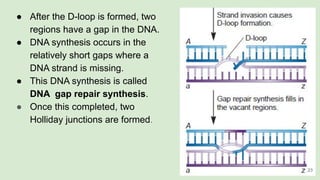
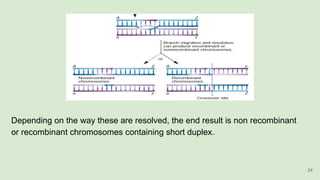
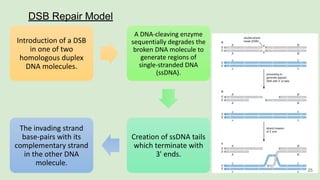
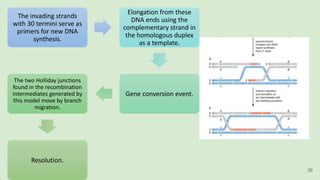
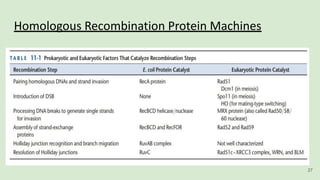
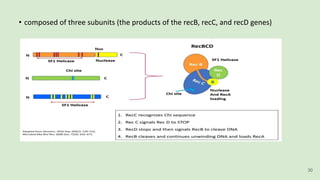
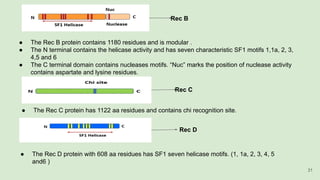
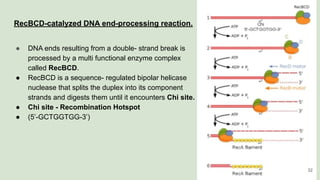
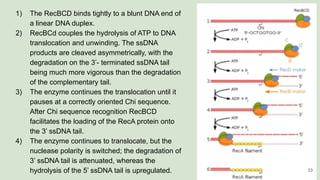
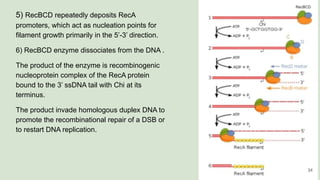
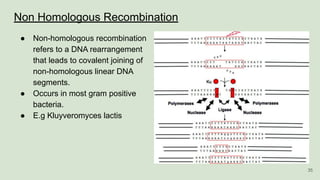
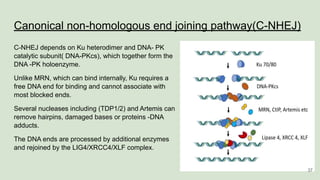
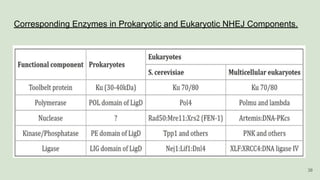
The document discusses recombination, specifically detailing DNA rearrangement and the formation of new gene combinations through mechanisms such as homologous, non-homologous, and site-specific recombination. It elaborates on various models of homologous recombination, including the Holliday model, Meselson-Radding model, and double-strand break model, outlining their processes and significance in genetic variation and DNA repair. Additionally, it describes the roles of proteins involved in homologous and non-homologous recombination, emphasizing their functions in maintaining genome integrity.