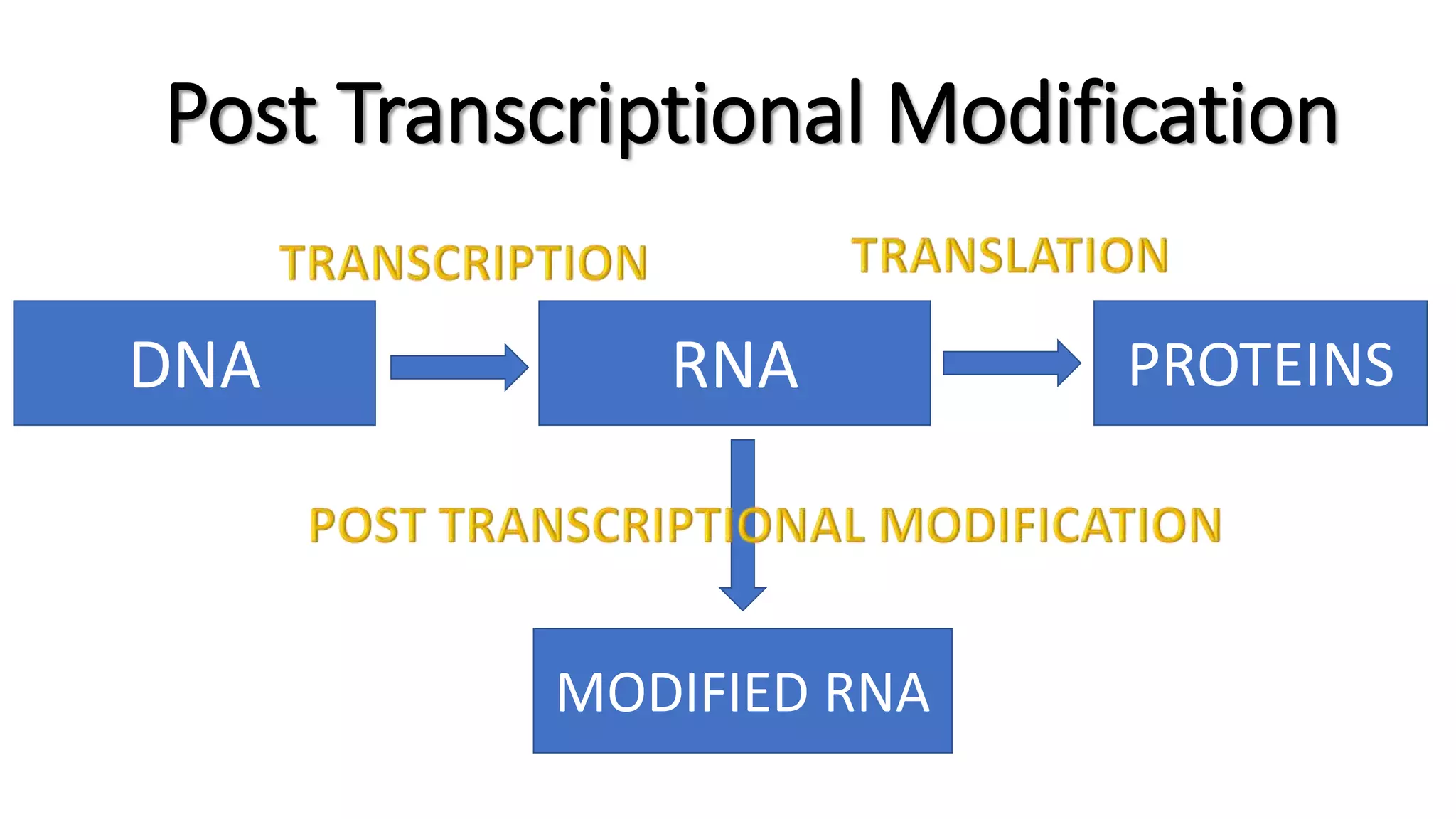
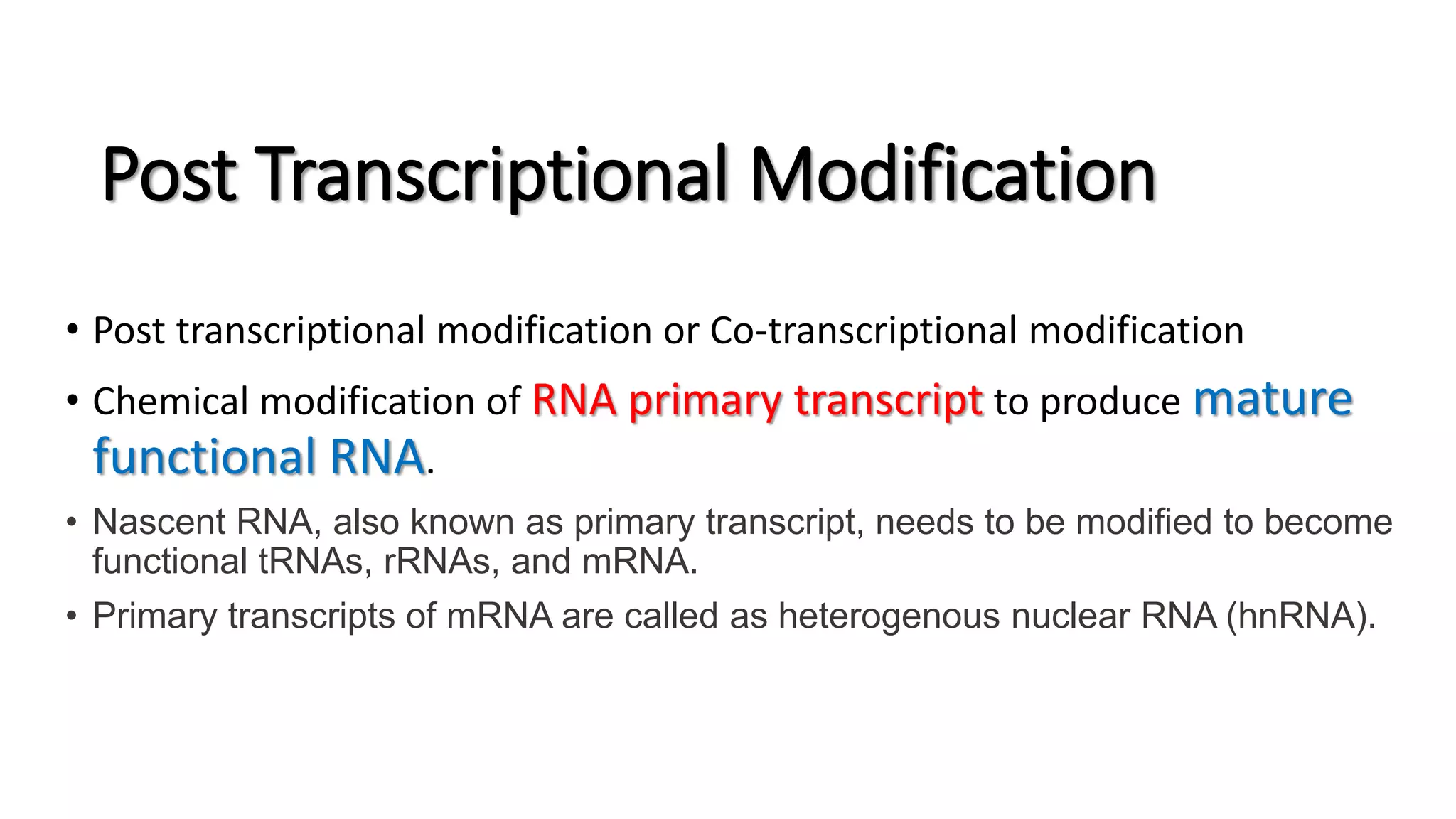
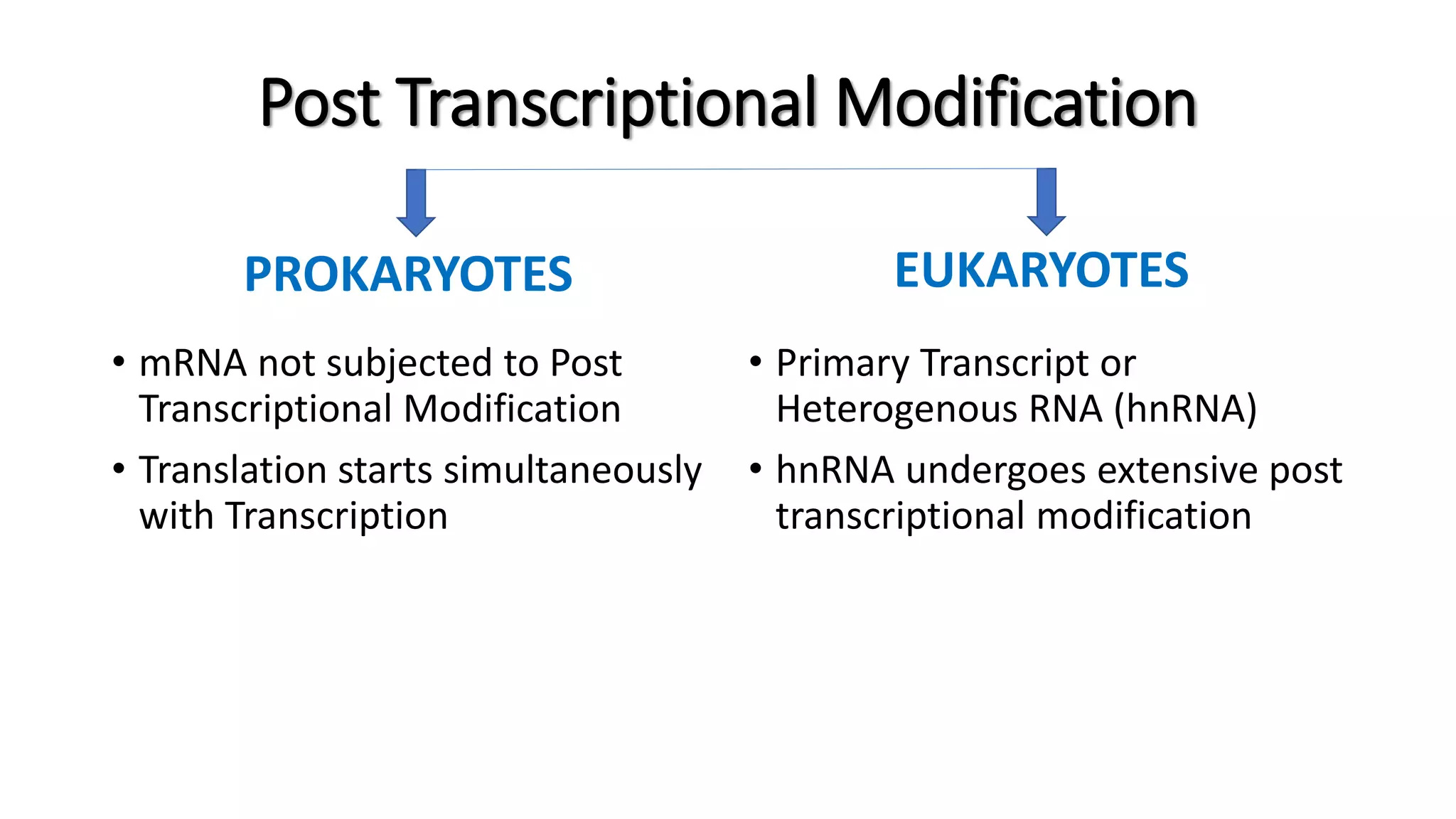
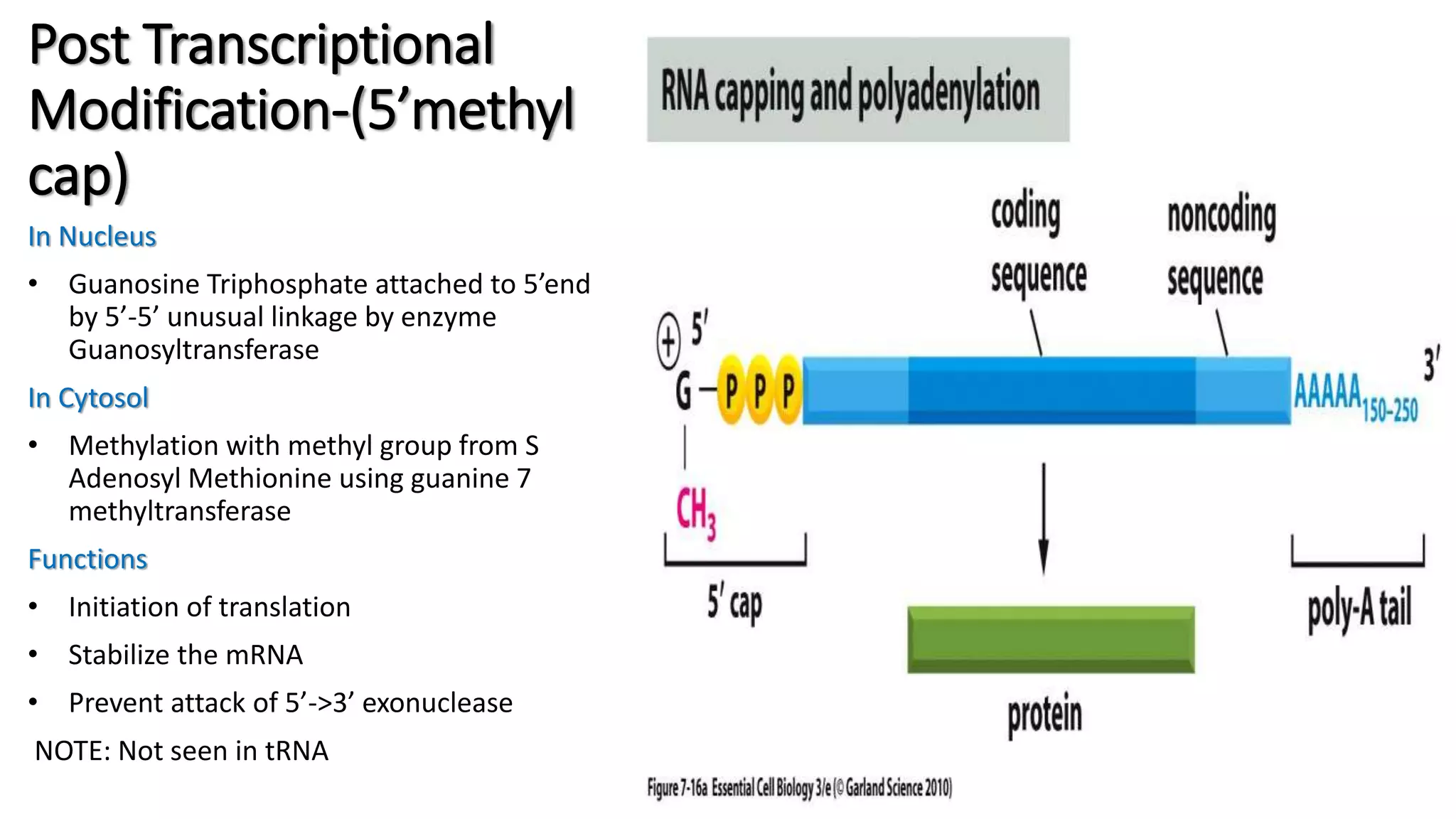
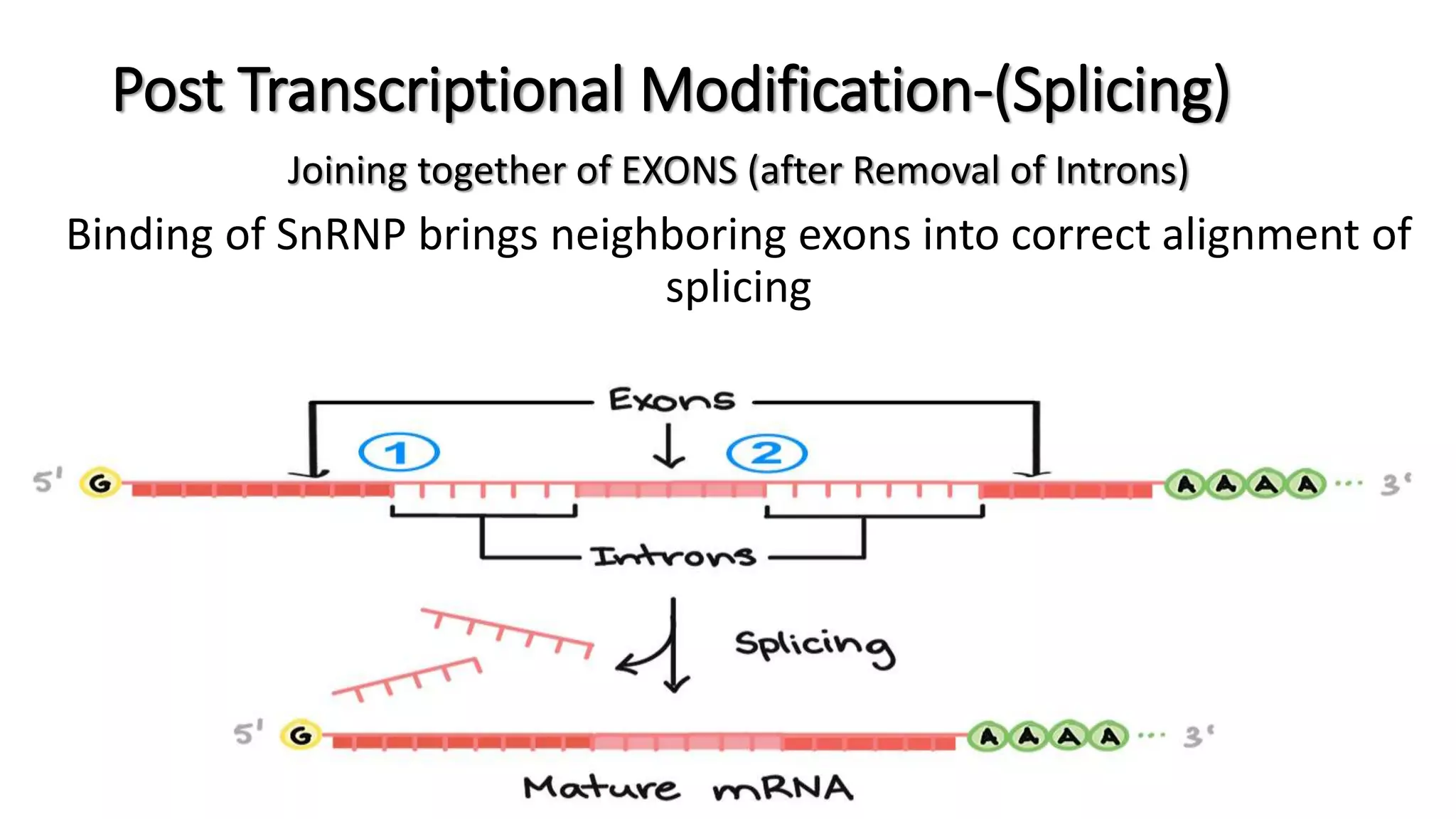
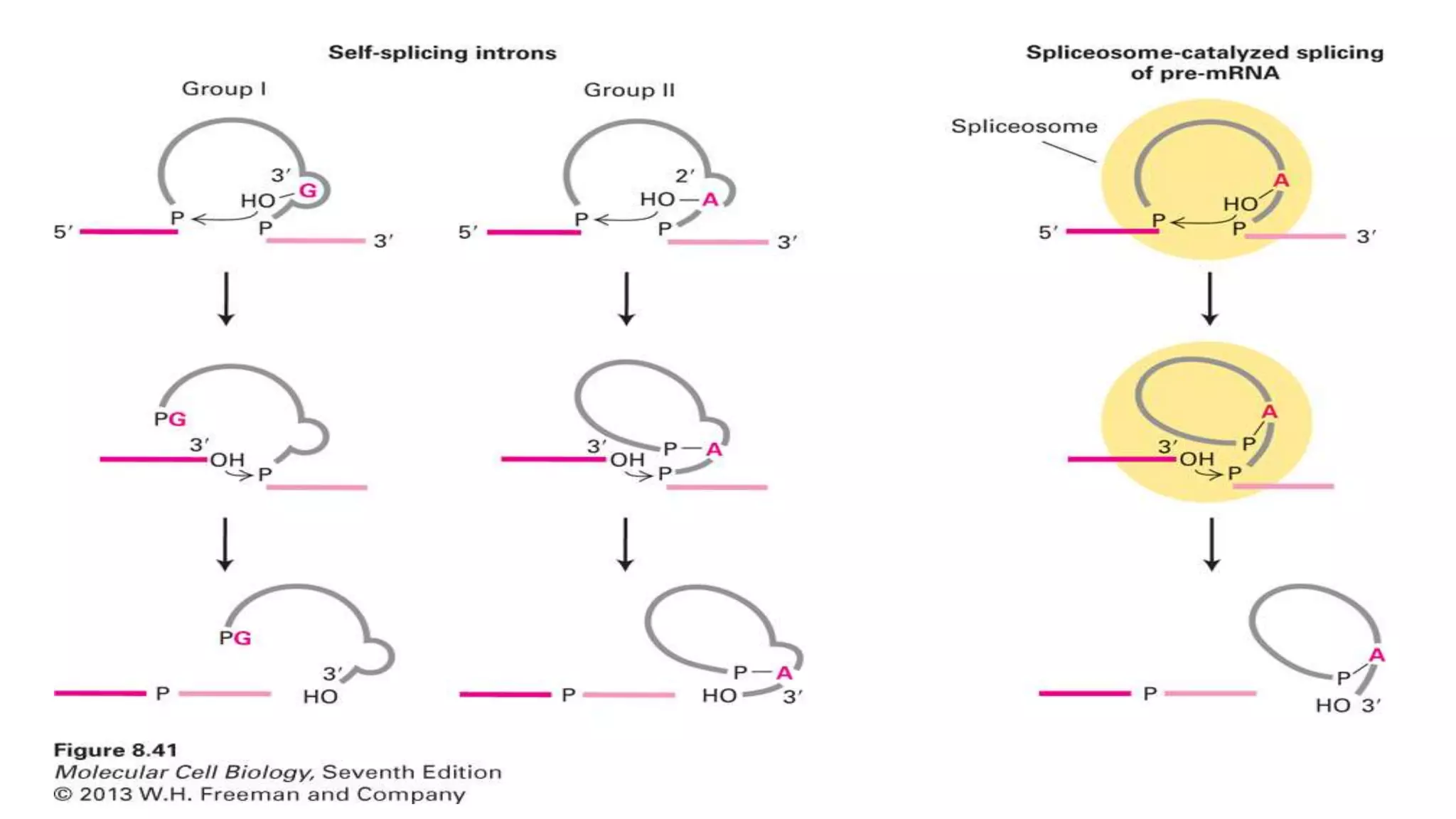
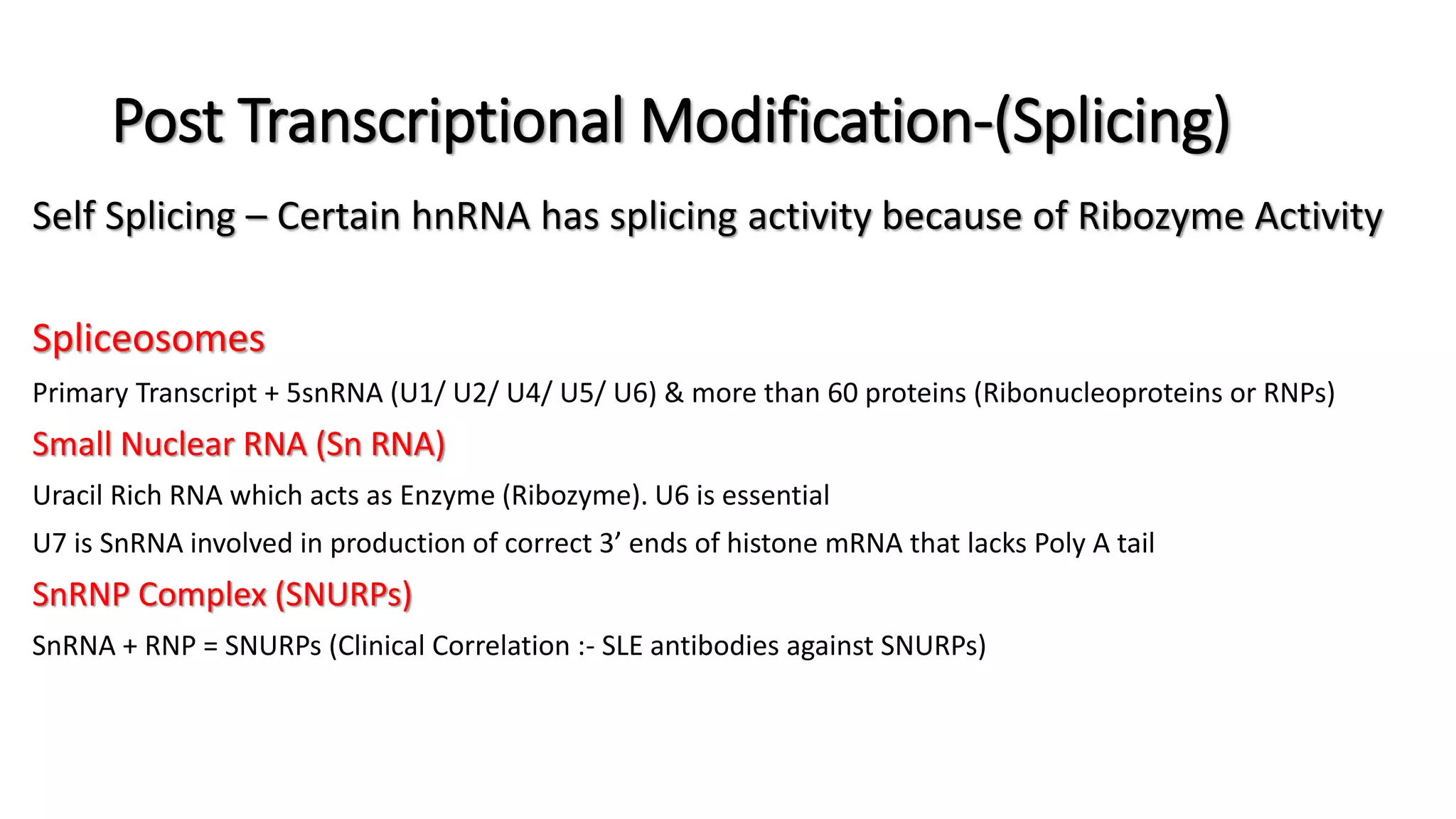
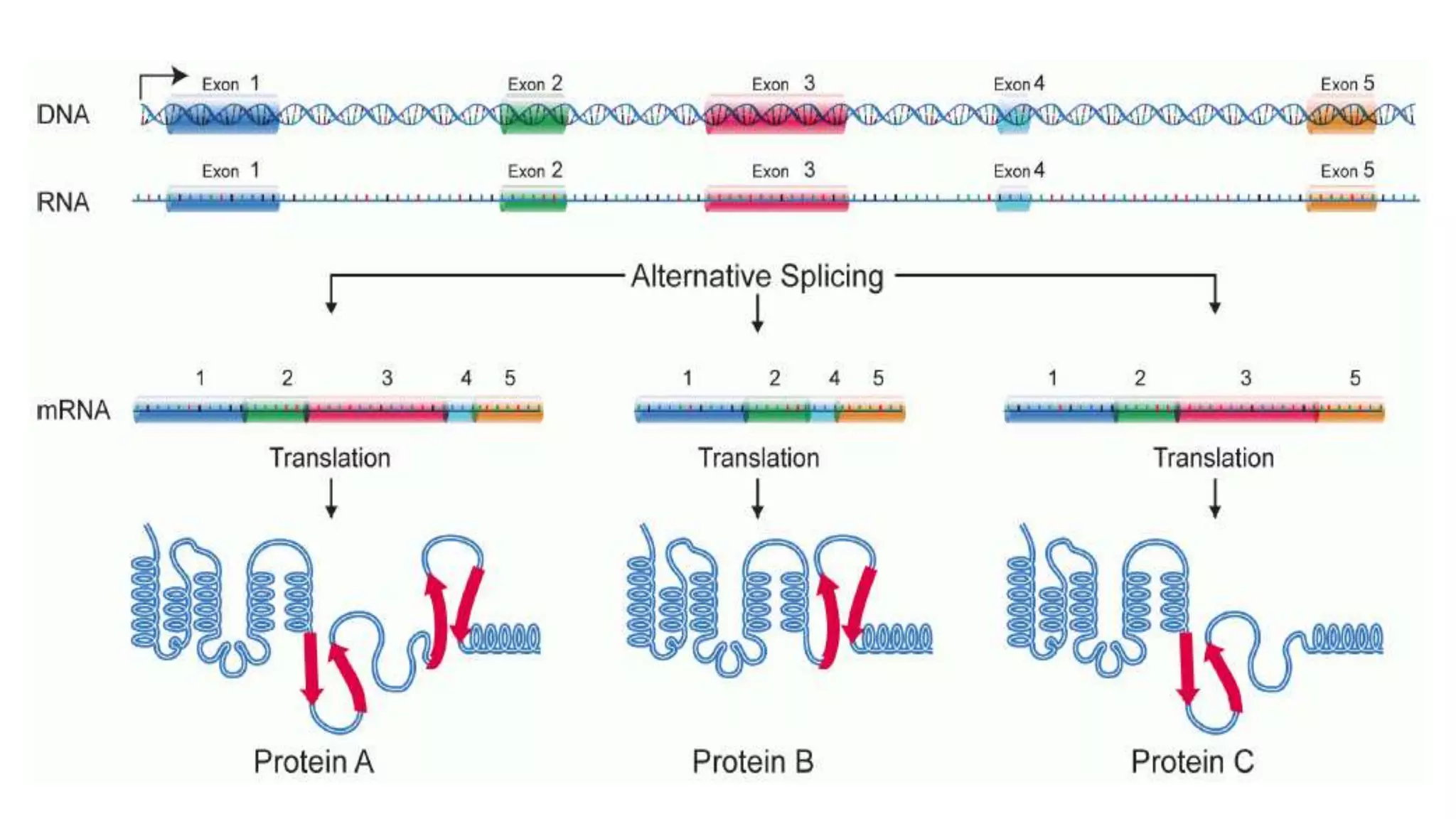
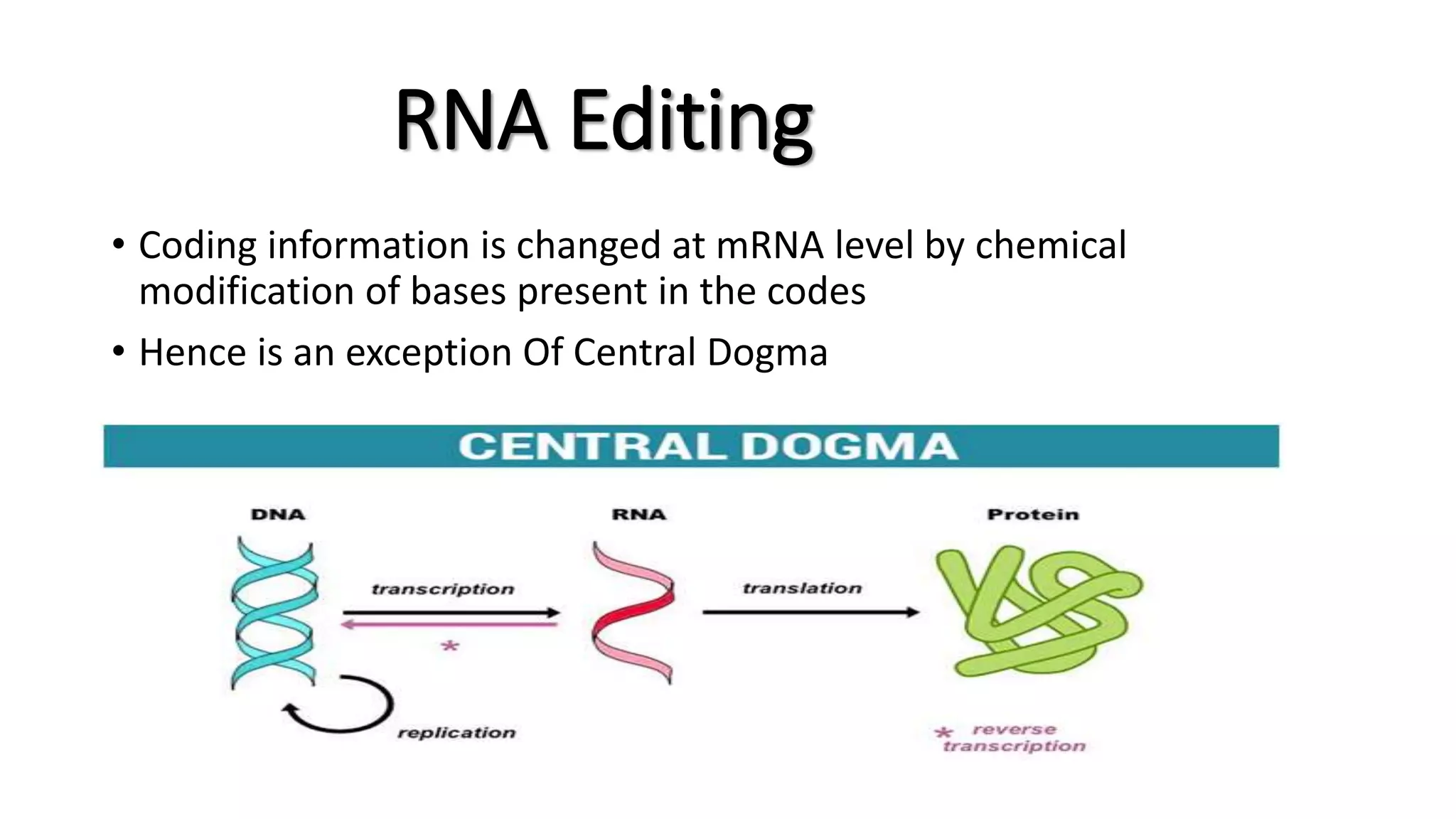
The document discusses post-transcriptional modifications in molecular biology, highlighting its significance in transforming nascent RNA into mature functional RNAs, specifically focusing on mRNA. It details the types of modifications that occur in eukaryotes, such as 5’ capping, polyadenylation, splicing, and alternative RNA processing. The document also emphasizes the roles and mechanisms of these modifications in ensuring mRNA stability and initiation of translation.