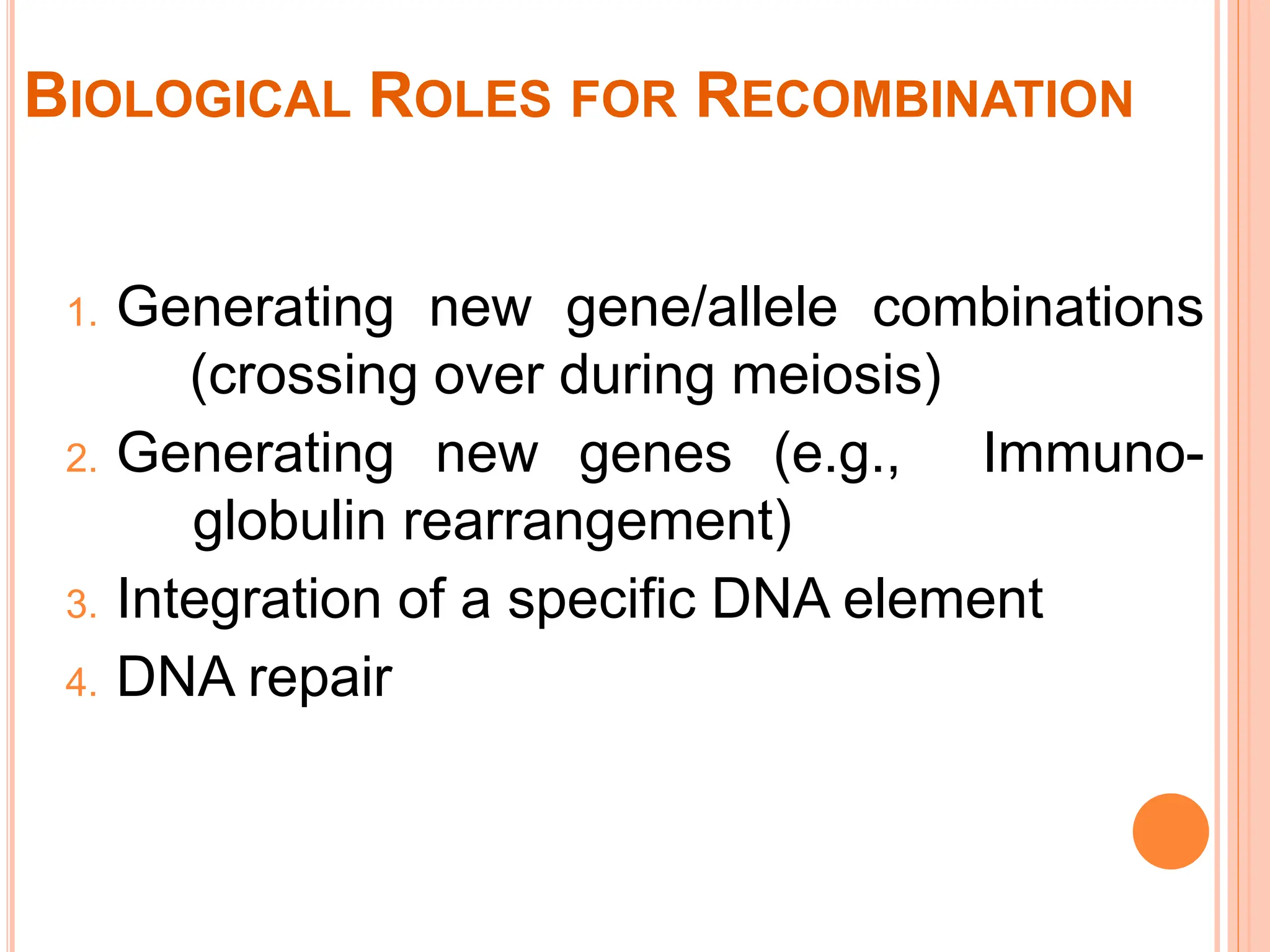
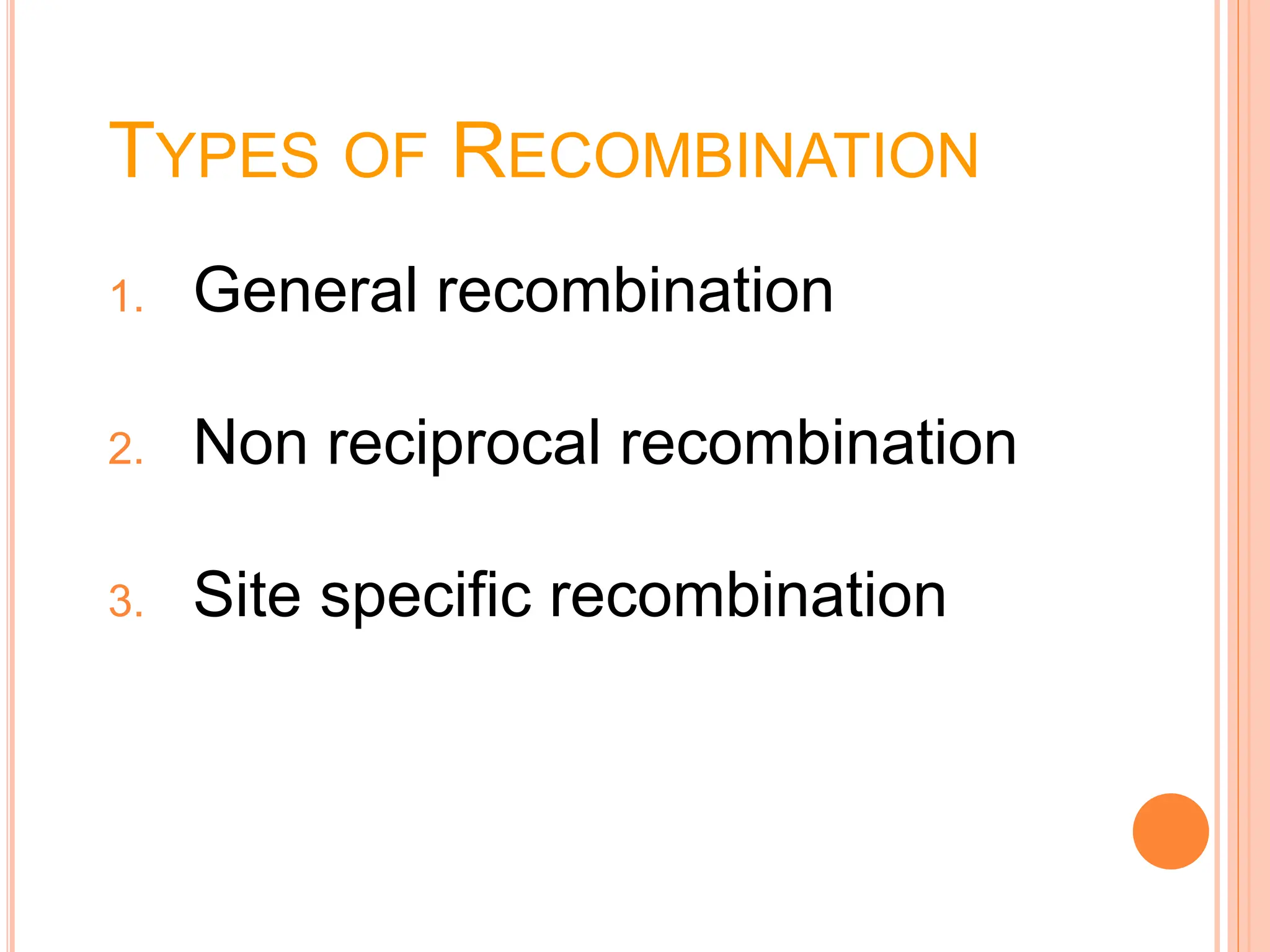
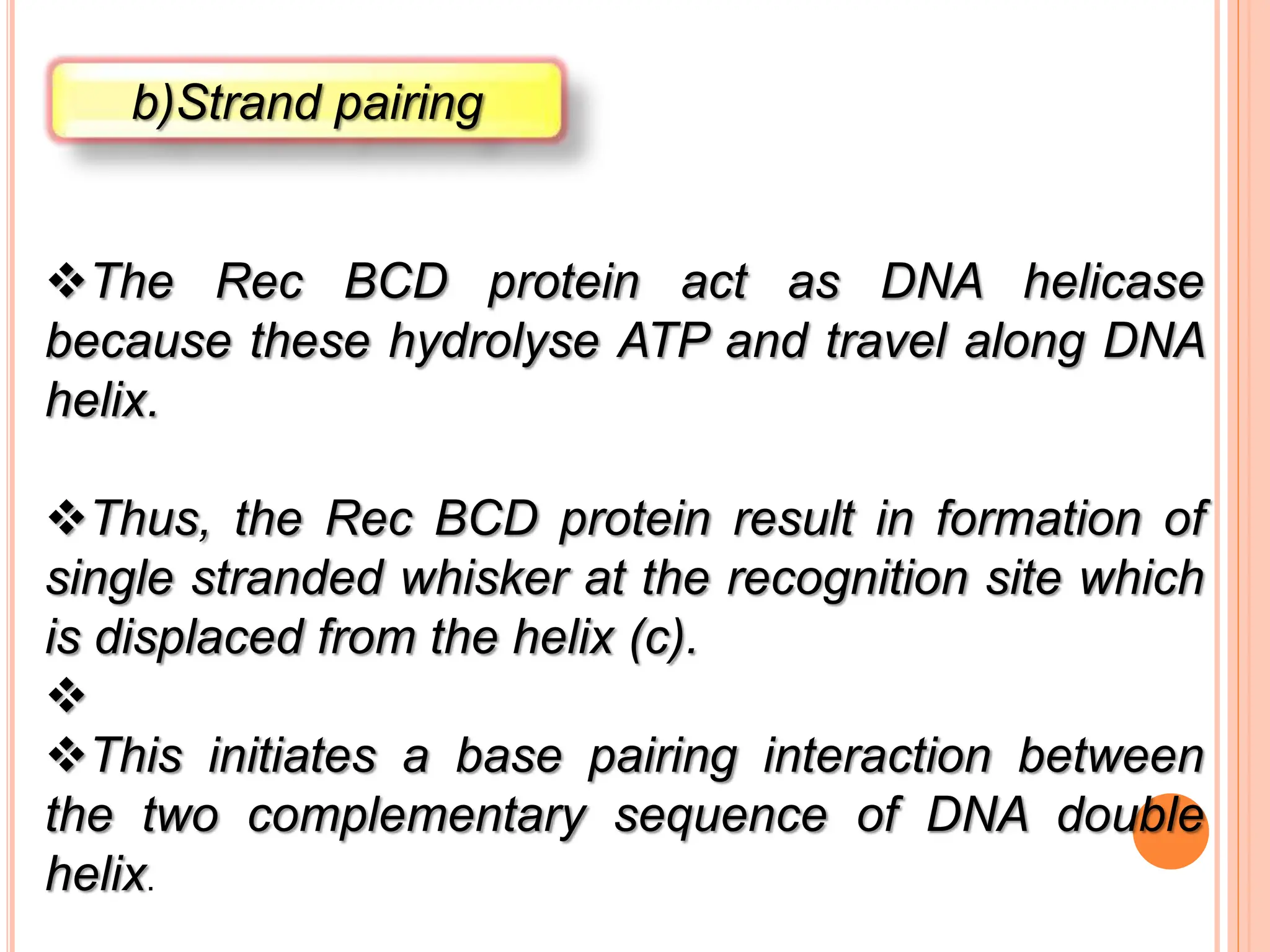
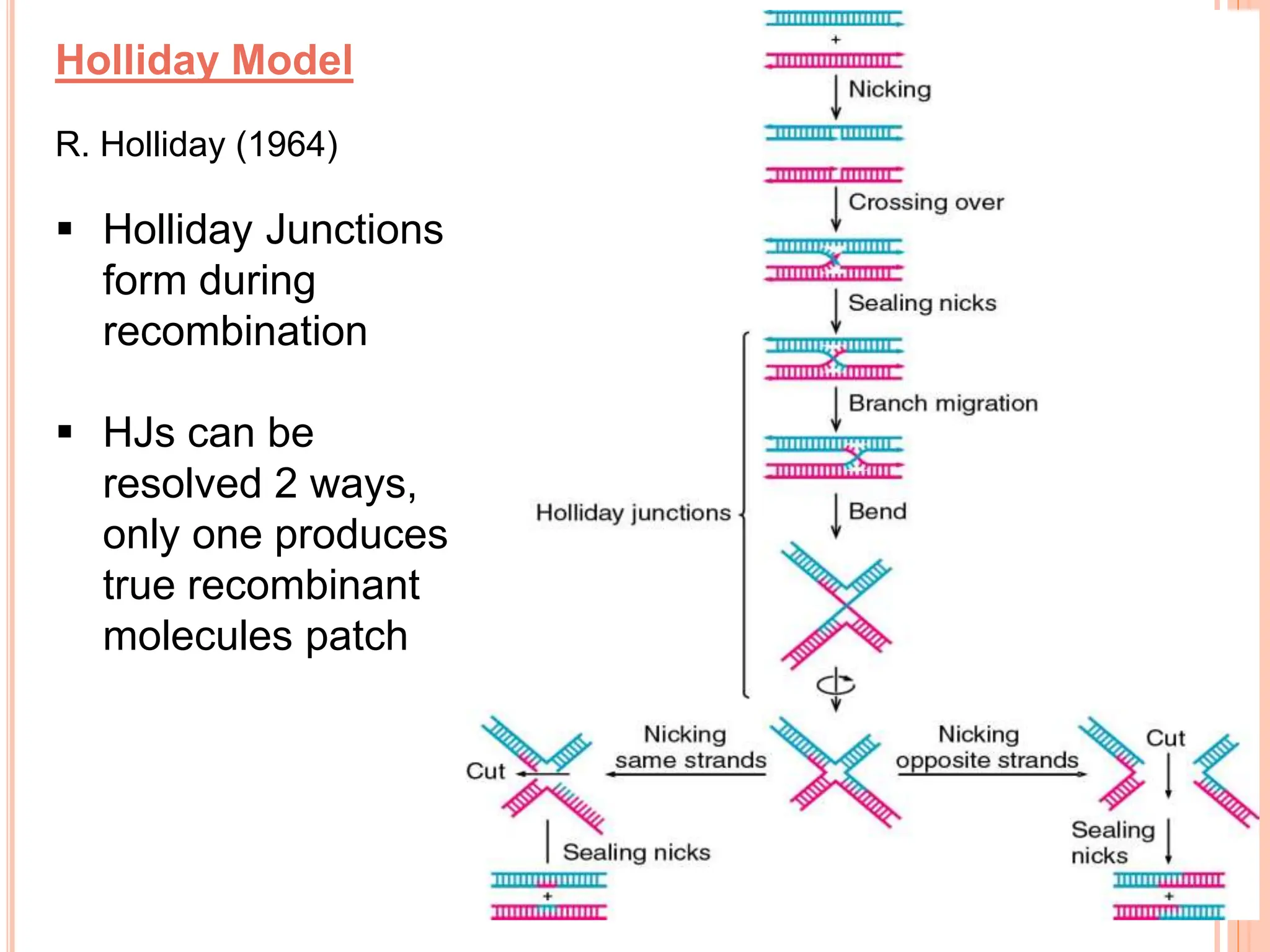
The document discusses genetic recombination, highlighting its biological roles, mechanisms, and types. Key biological roles include generating gene combinations and DNA repair, while mechanisms involve breakage and reunion, and copying processes. Types of recombination covered are general, non-reciprocal, and site-specific recombination, elaborating on their respective processes and models.