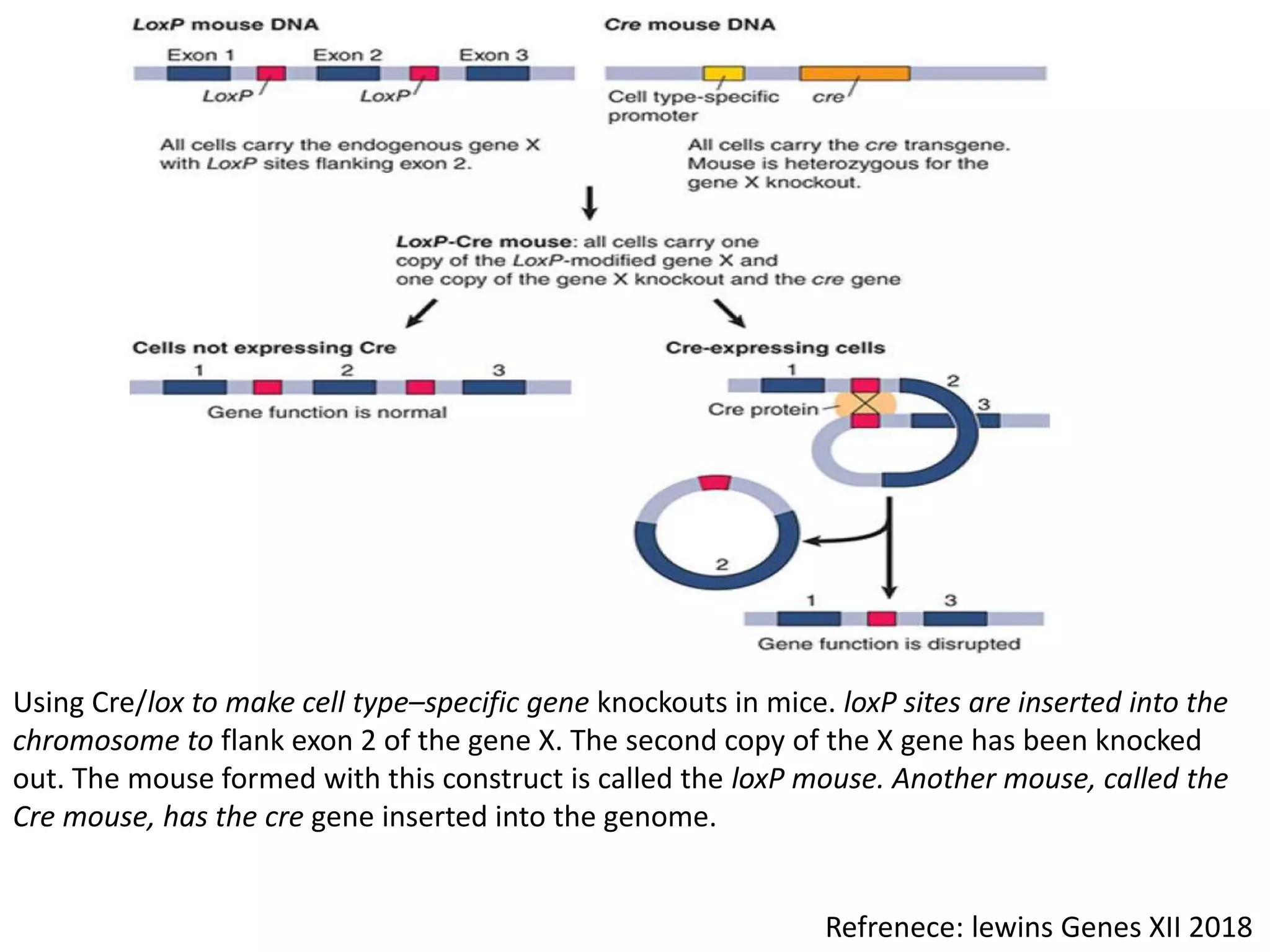
1. The document discusses models of homologous recombination including the Holliday model and the double-strand break repair model. It describes the key steps and proteins involved in each model.
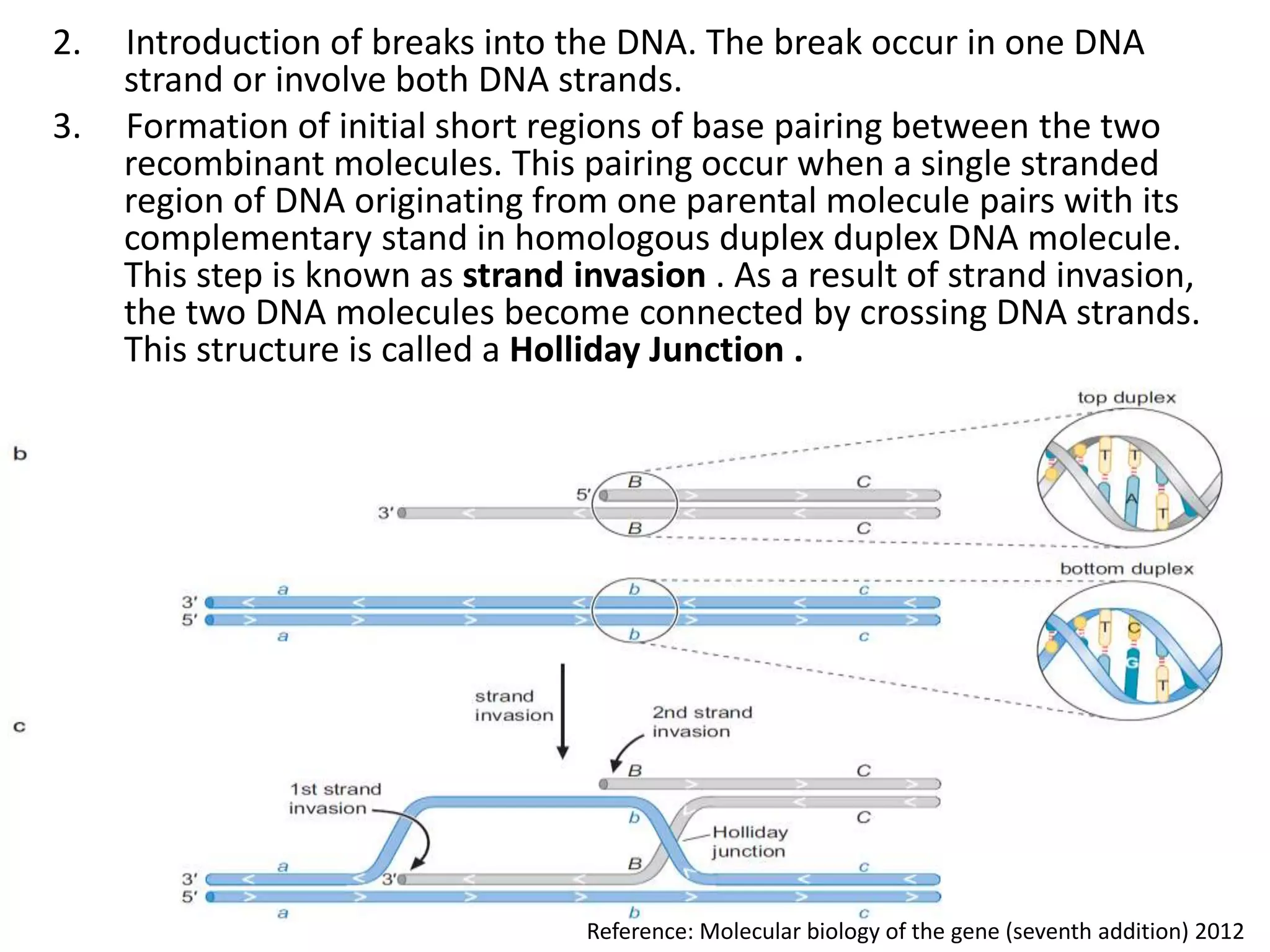
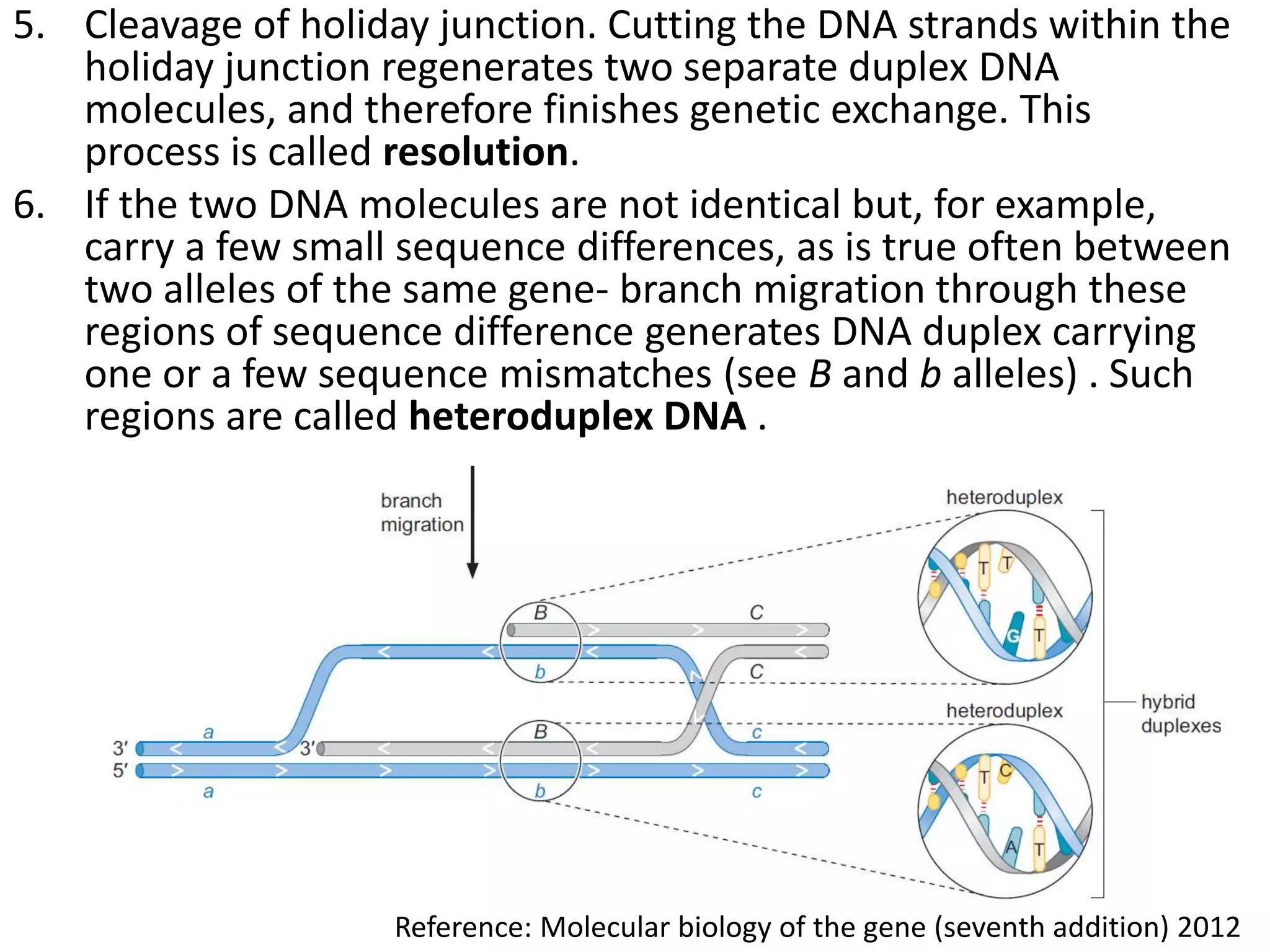
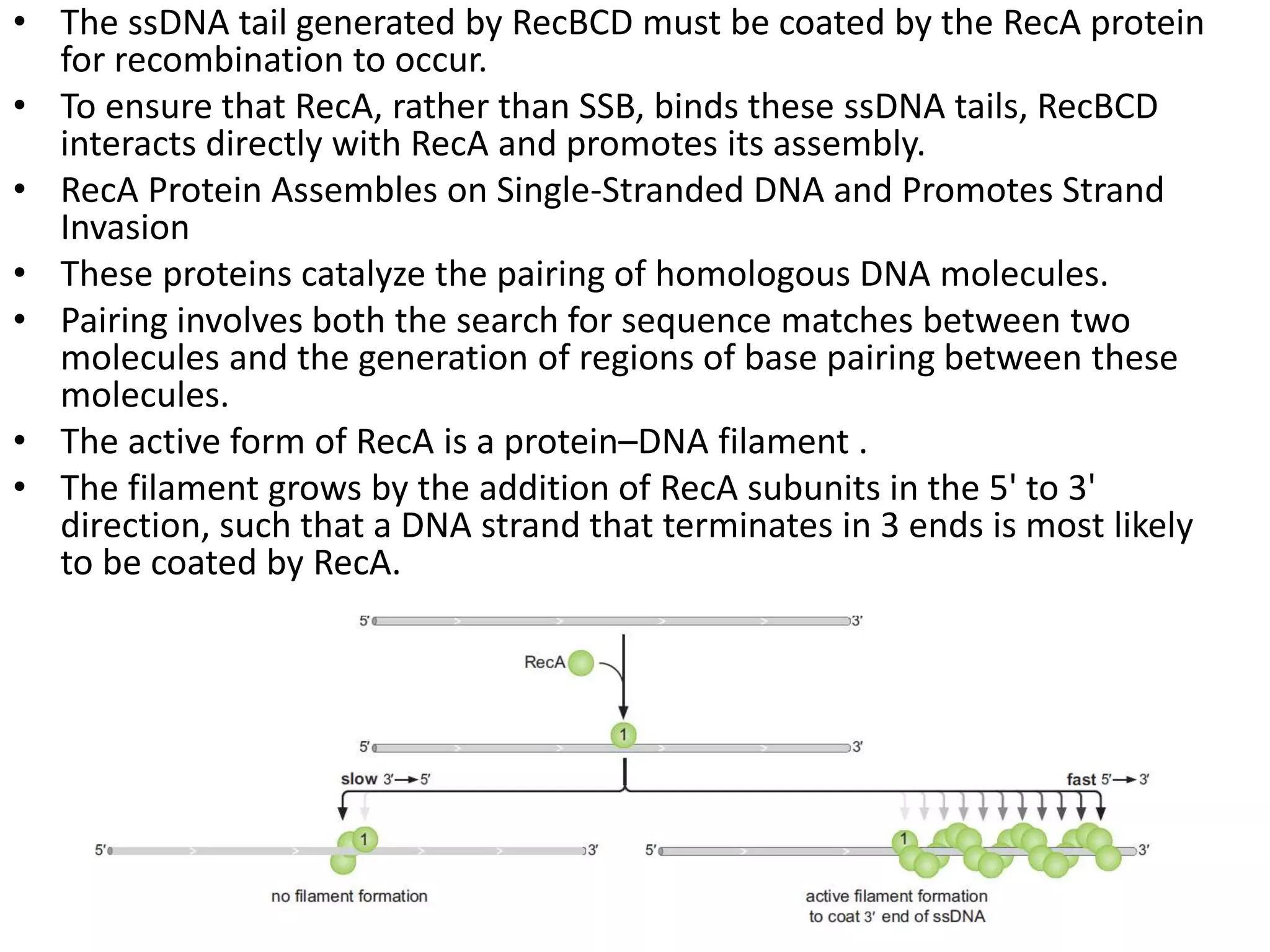
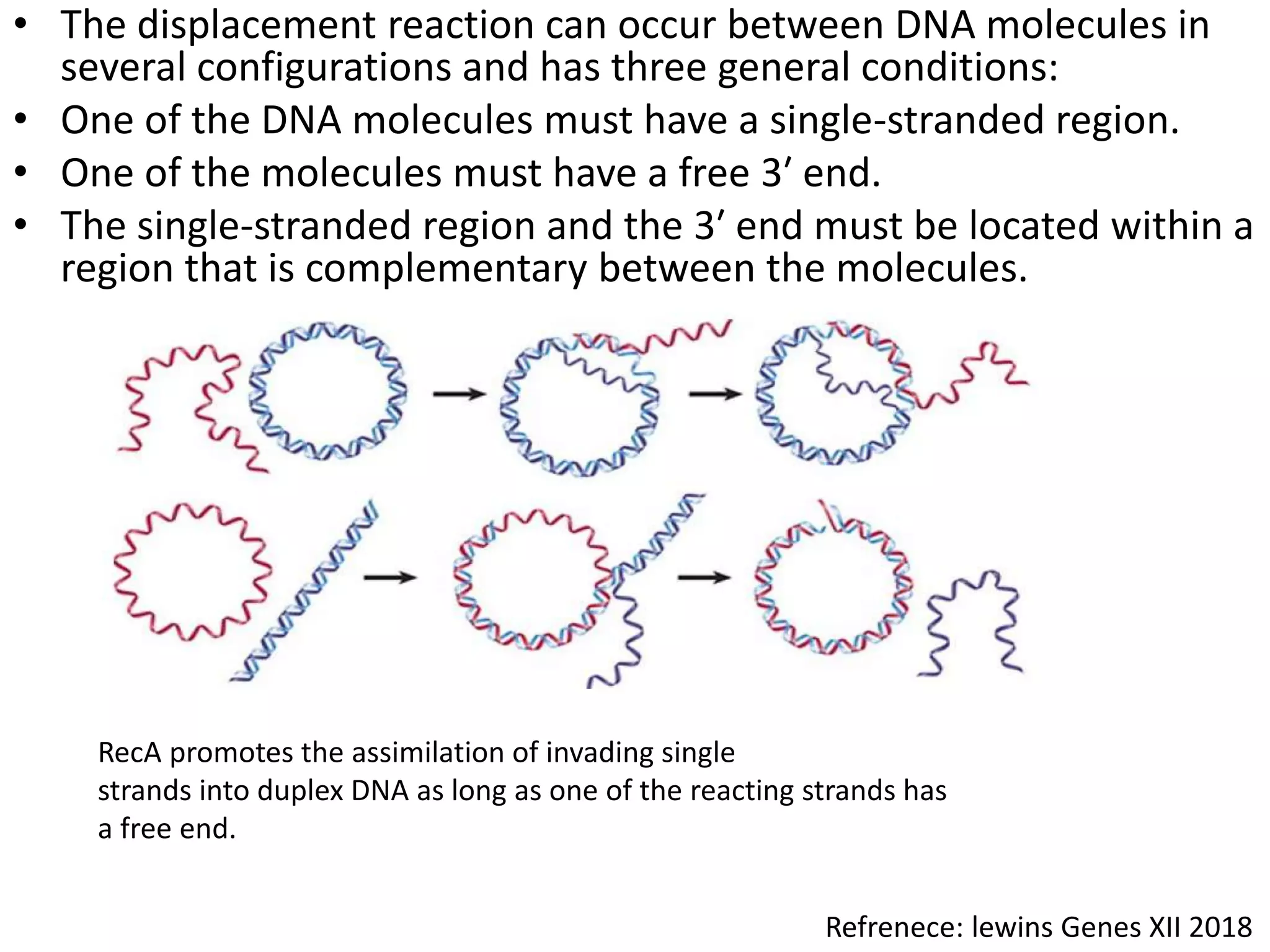
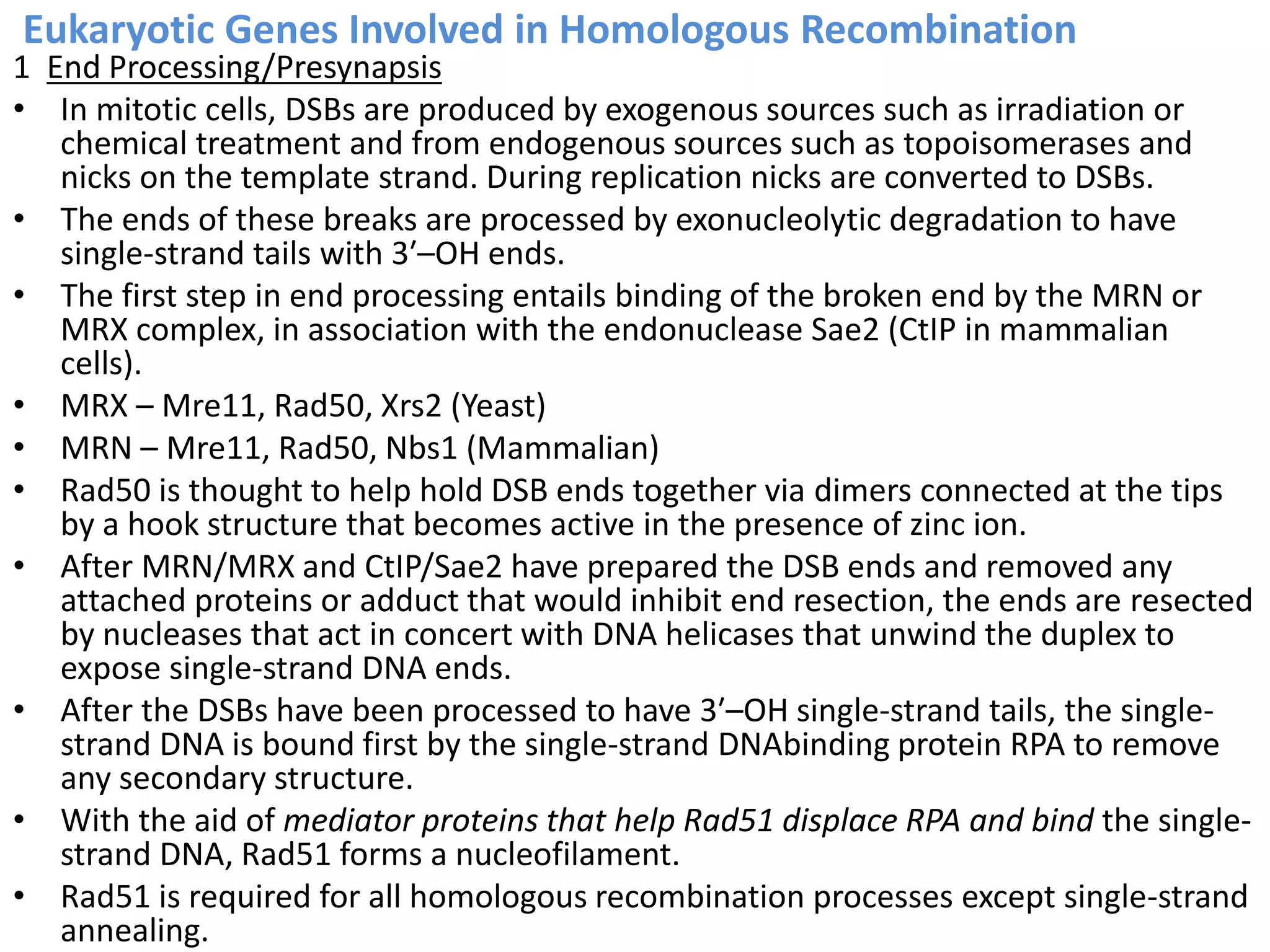
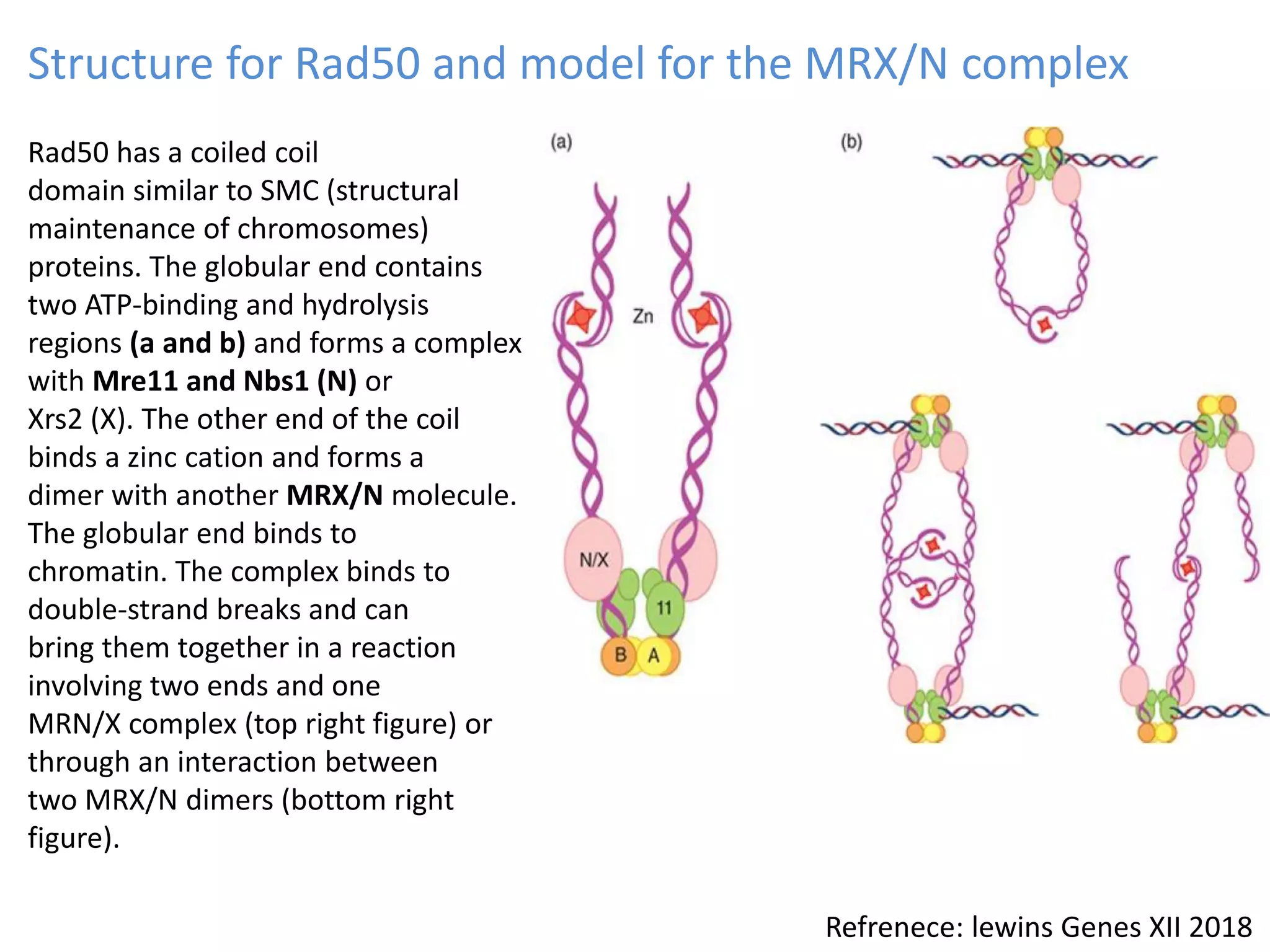
2. Recombination involves the breakage and rejoining of DNA. In eukaryotes, the MRN/X complex processes DNA breaks. The Rad51 and Rad54 proteins then facilitate strand invasion and D-loop formation during homologous pairing.
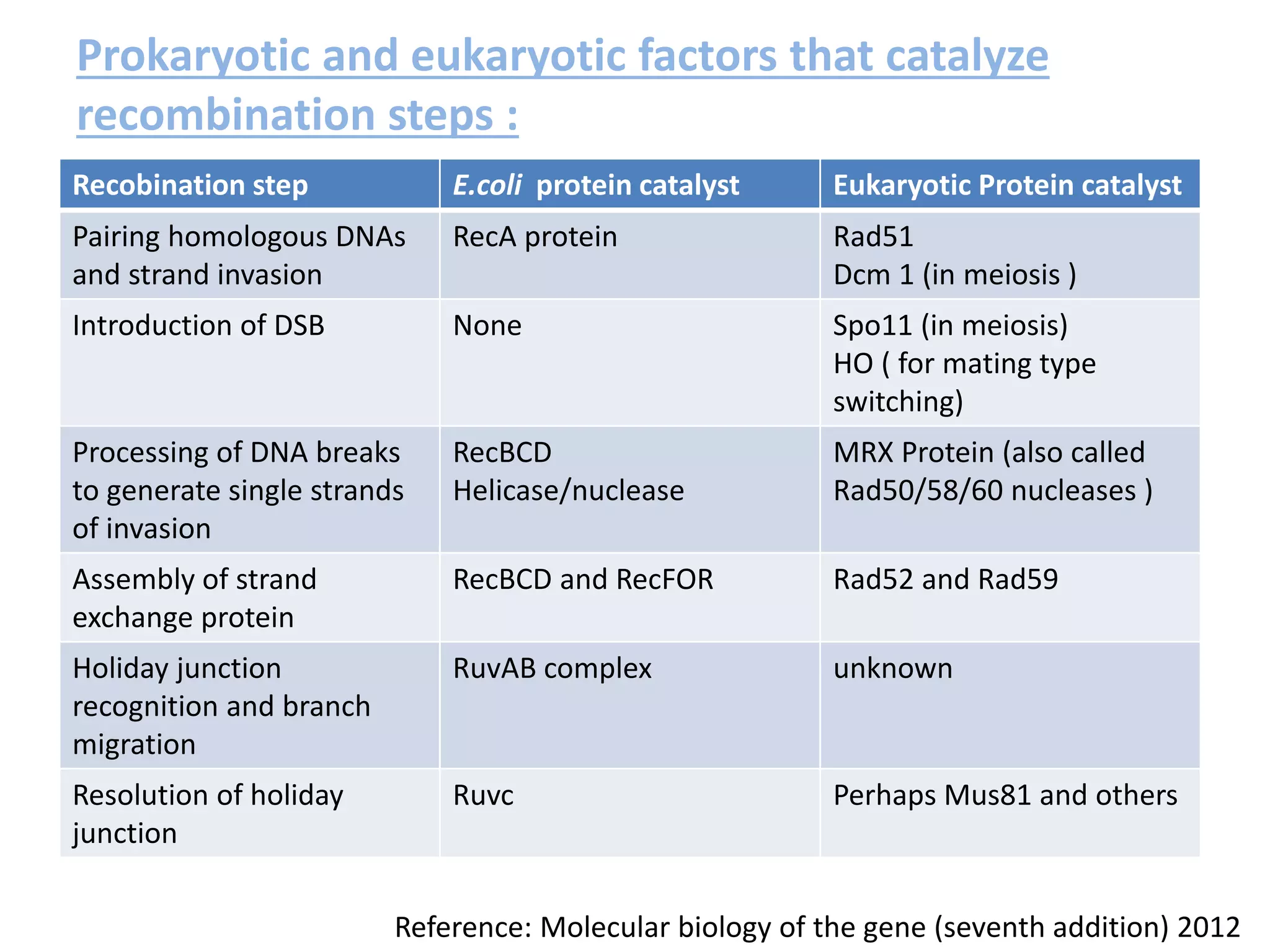
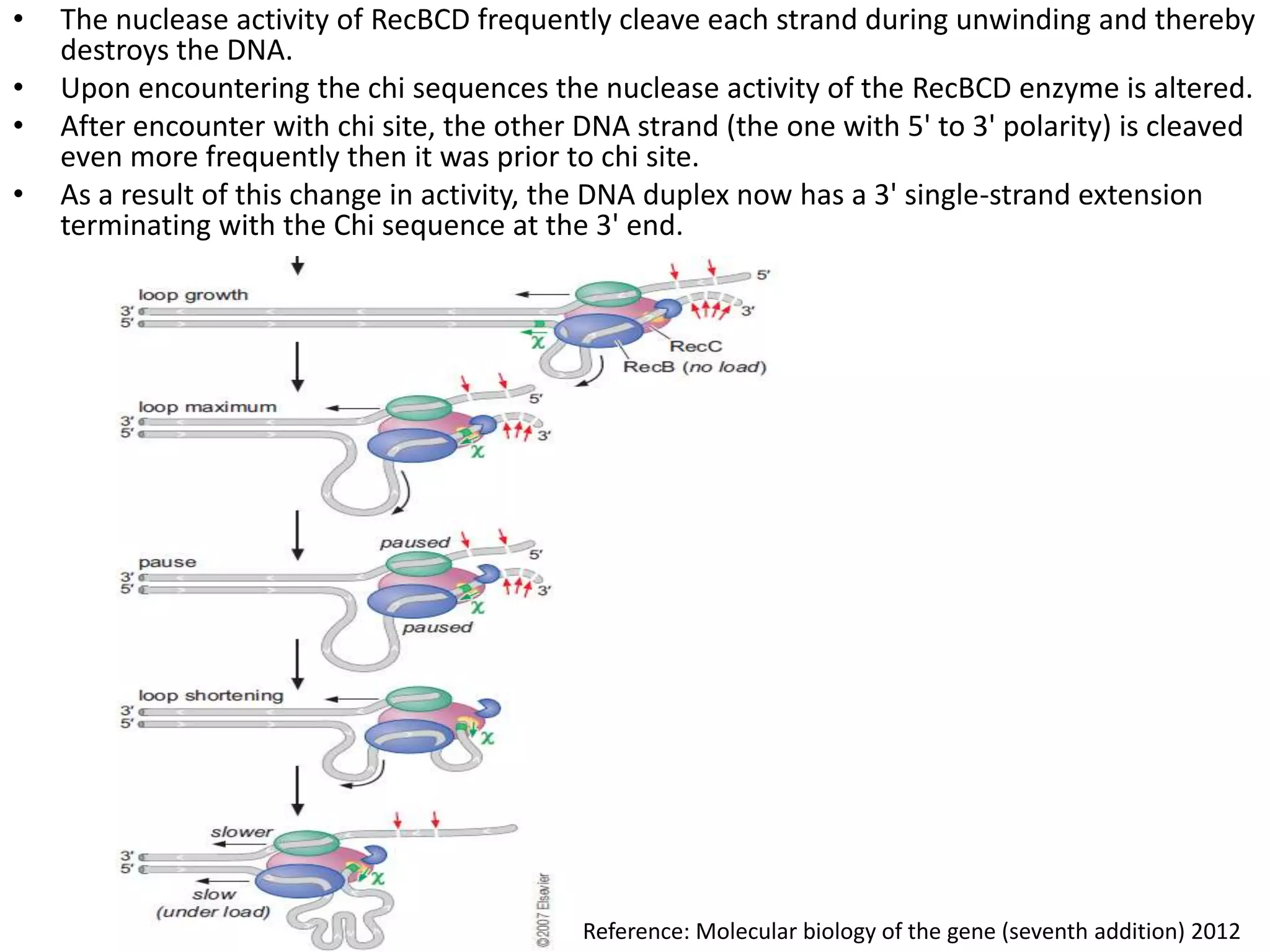
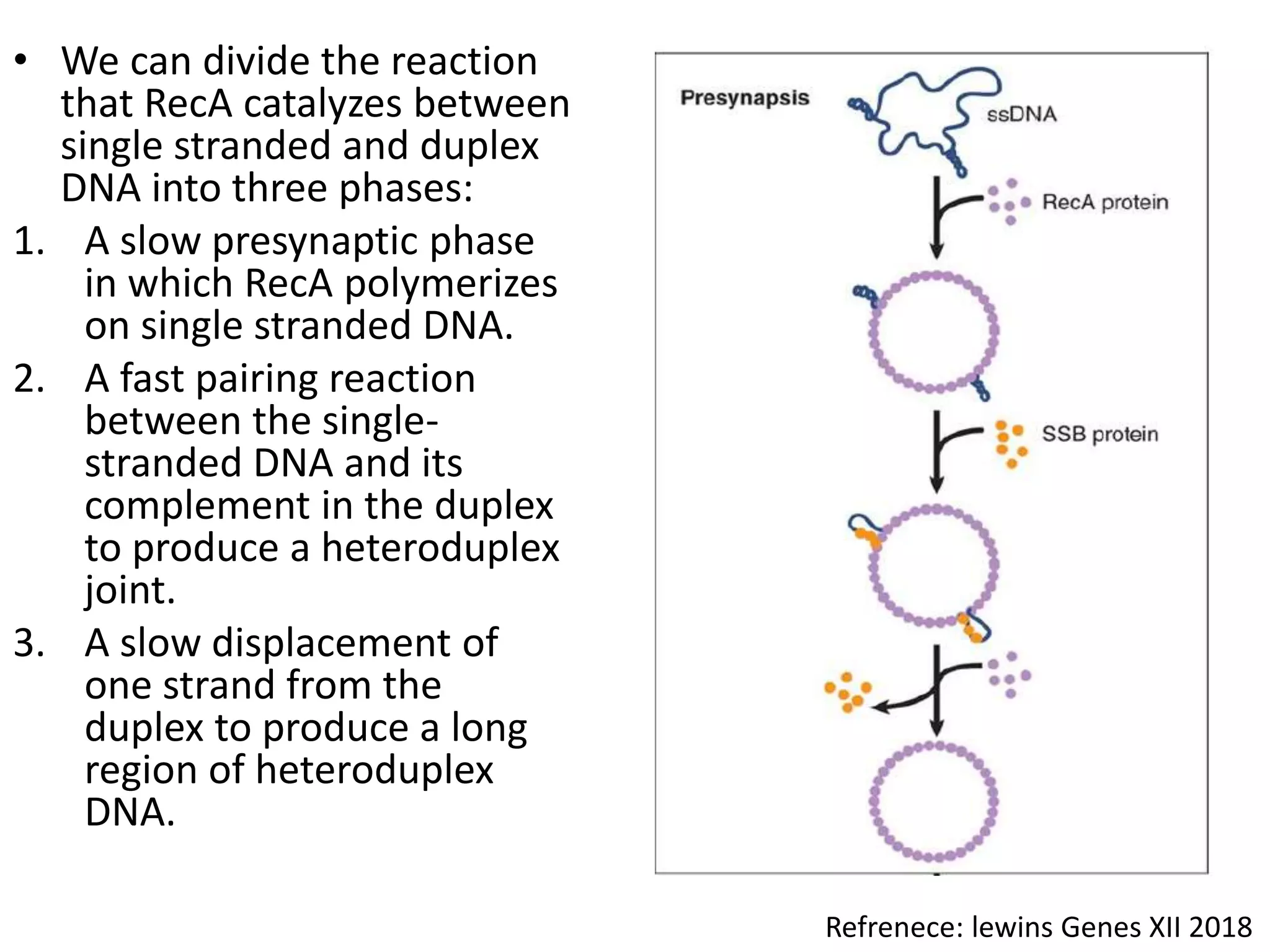
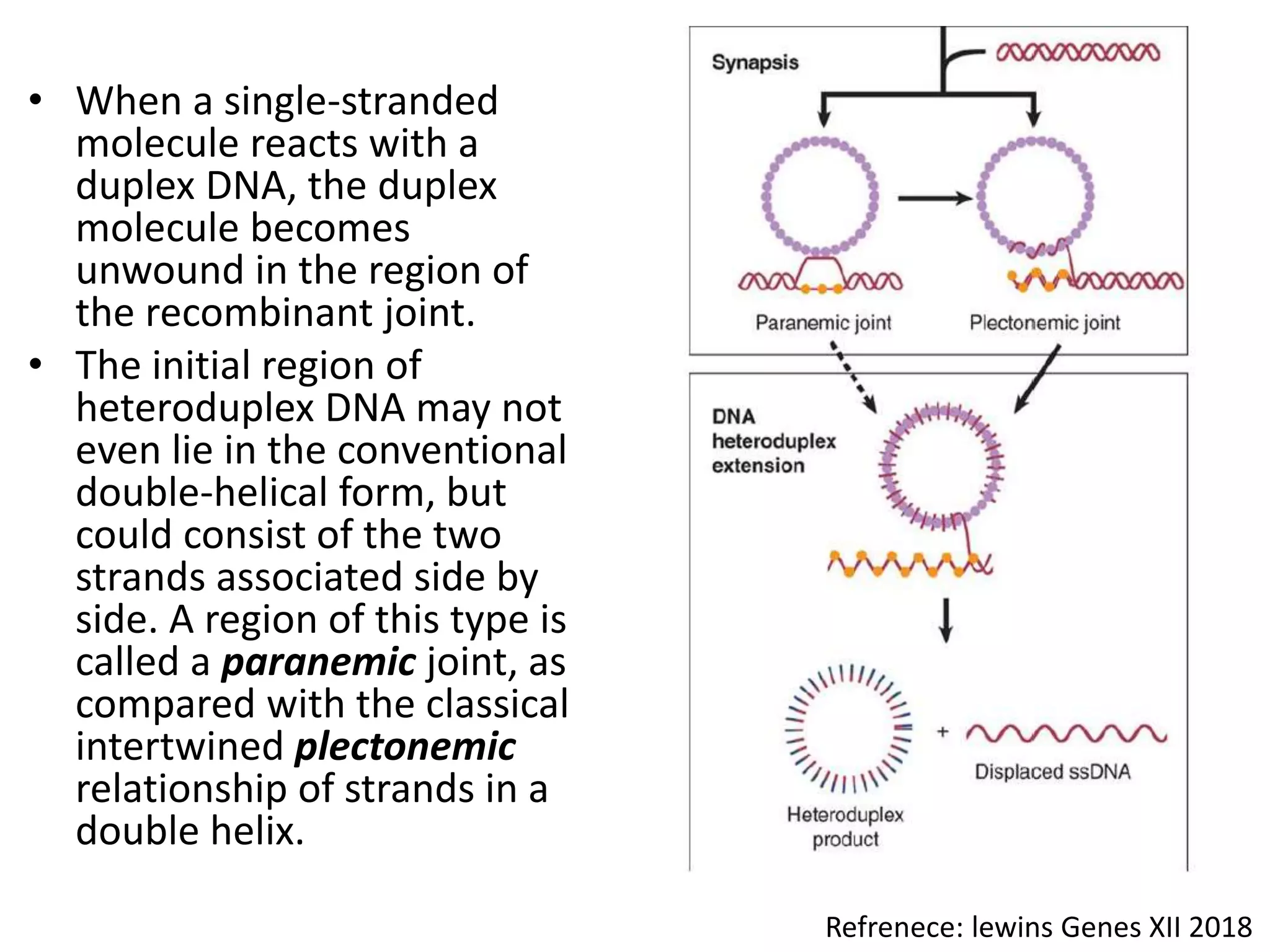
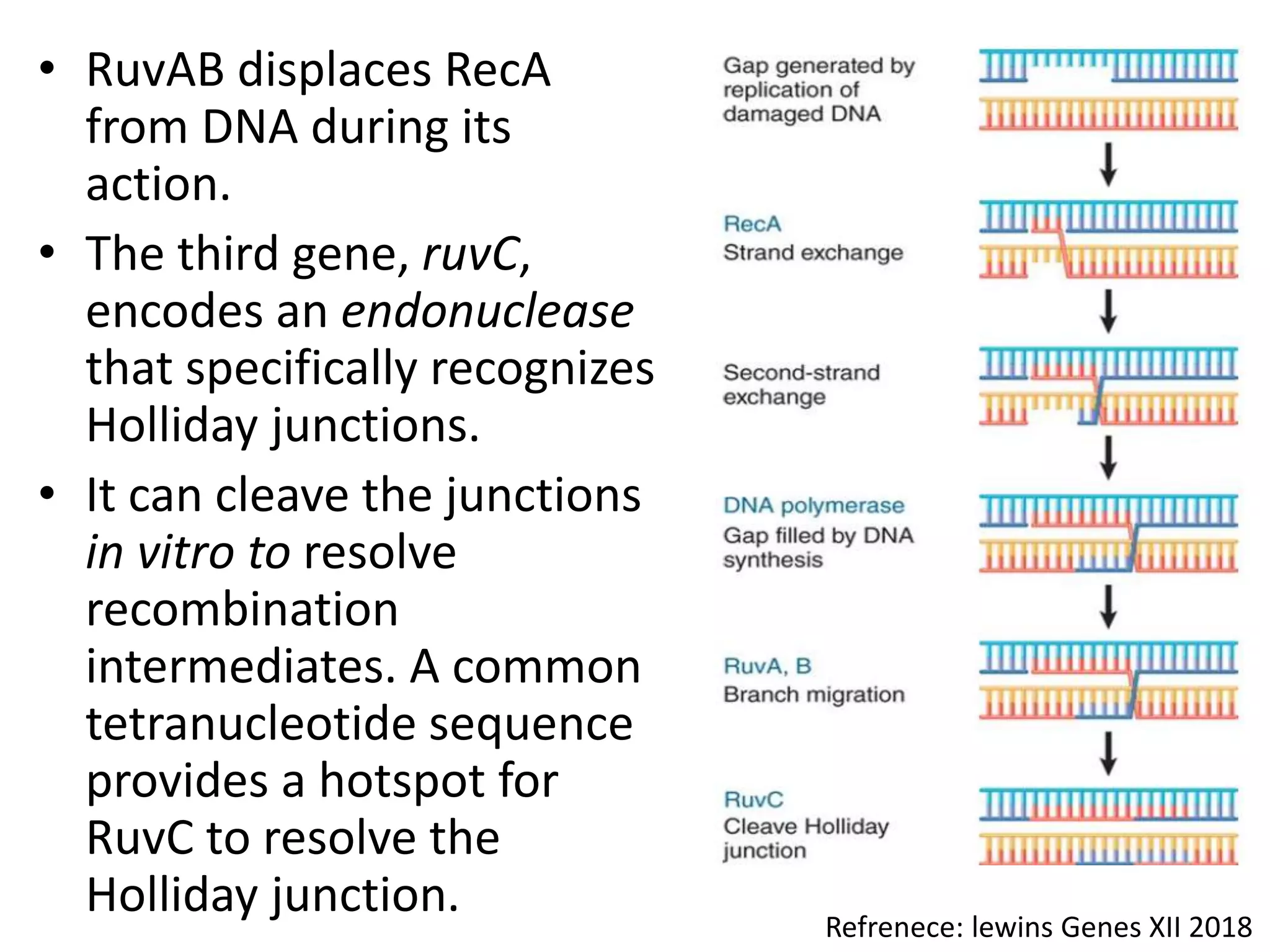
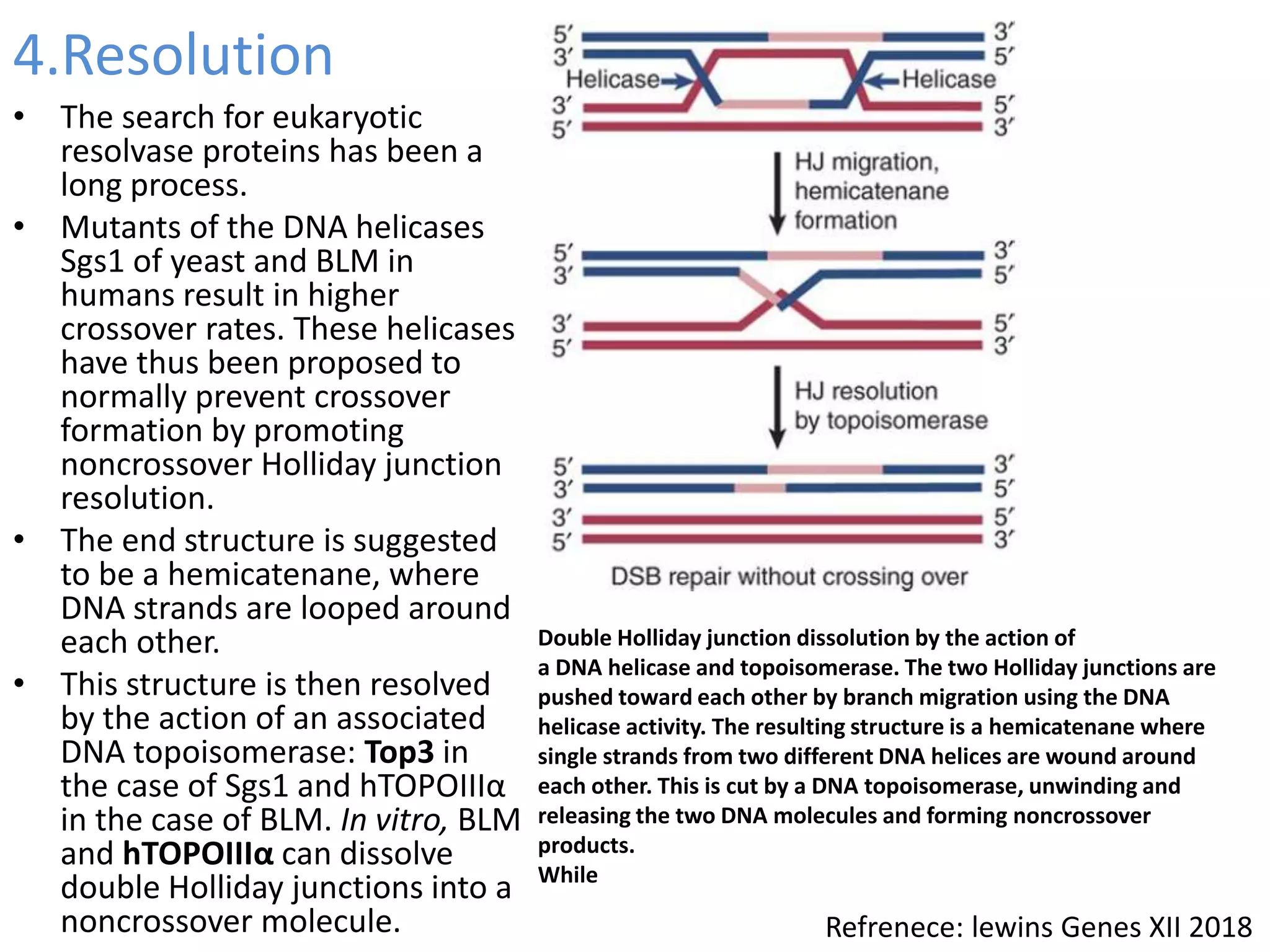
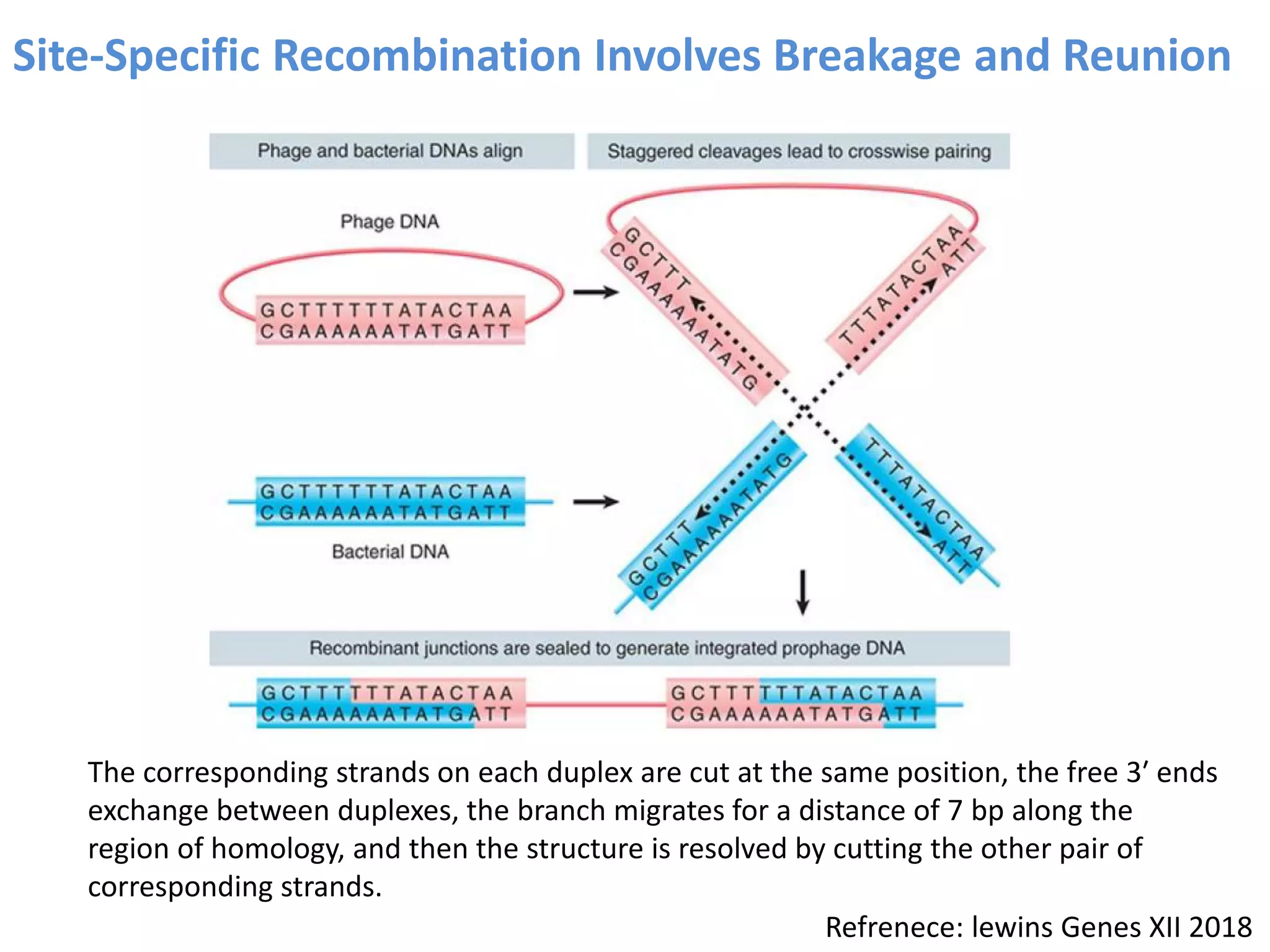
3. Homologous recombination proteins from bacteria and eukaryotes catalyze different steps of the process. In E. coli, RecBCD introduces breaks and generates single strands for RecA to perform strand exchange, while RuvAB and Ruv