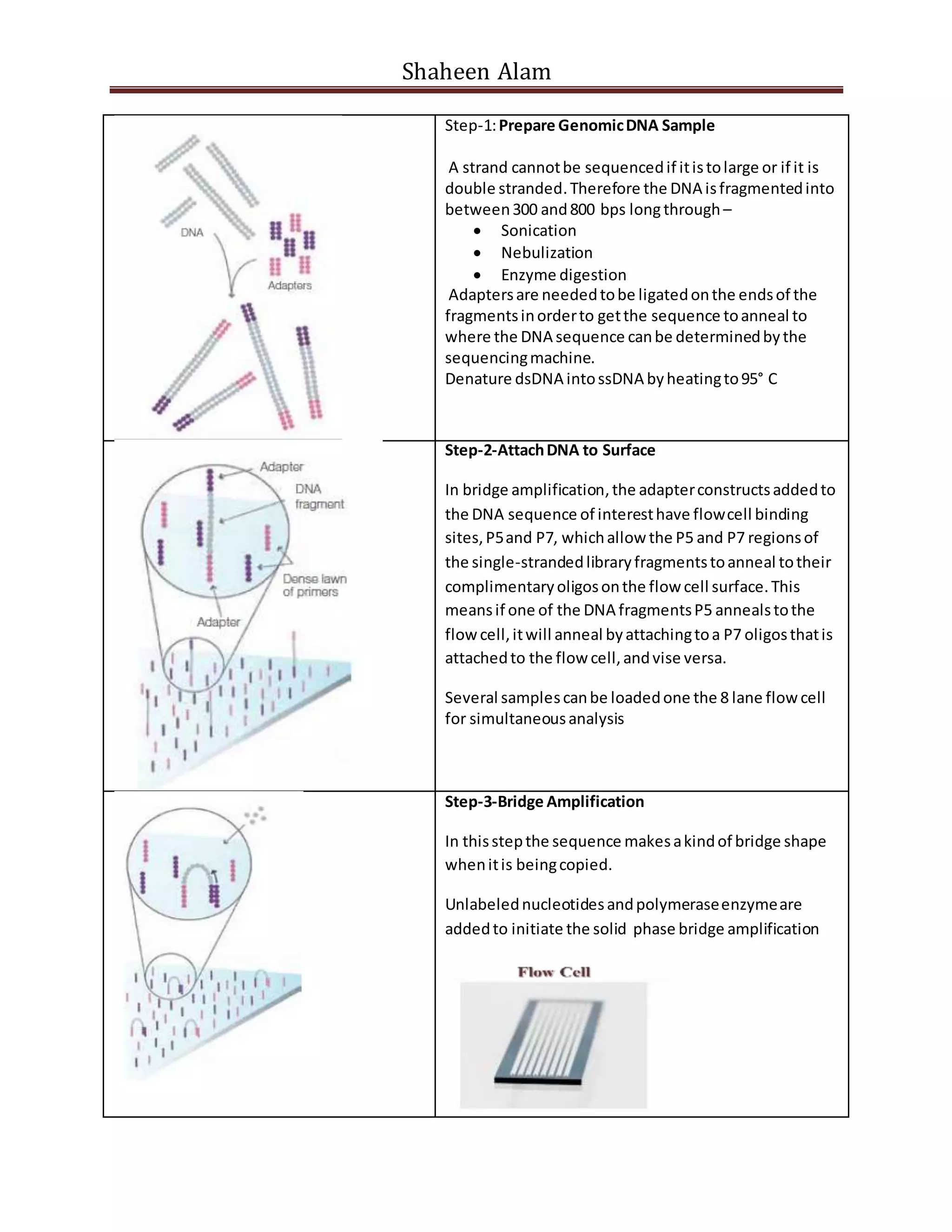
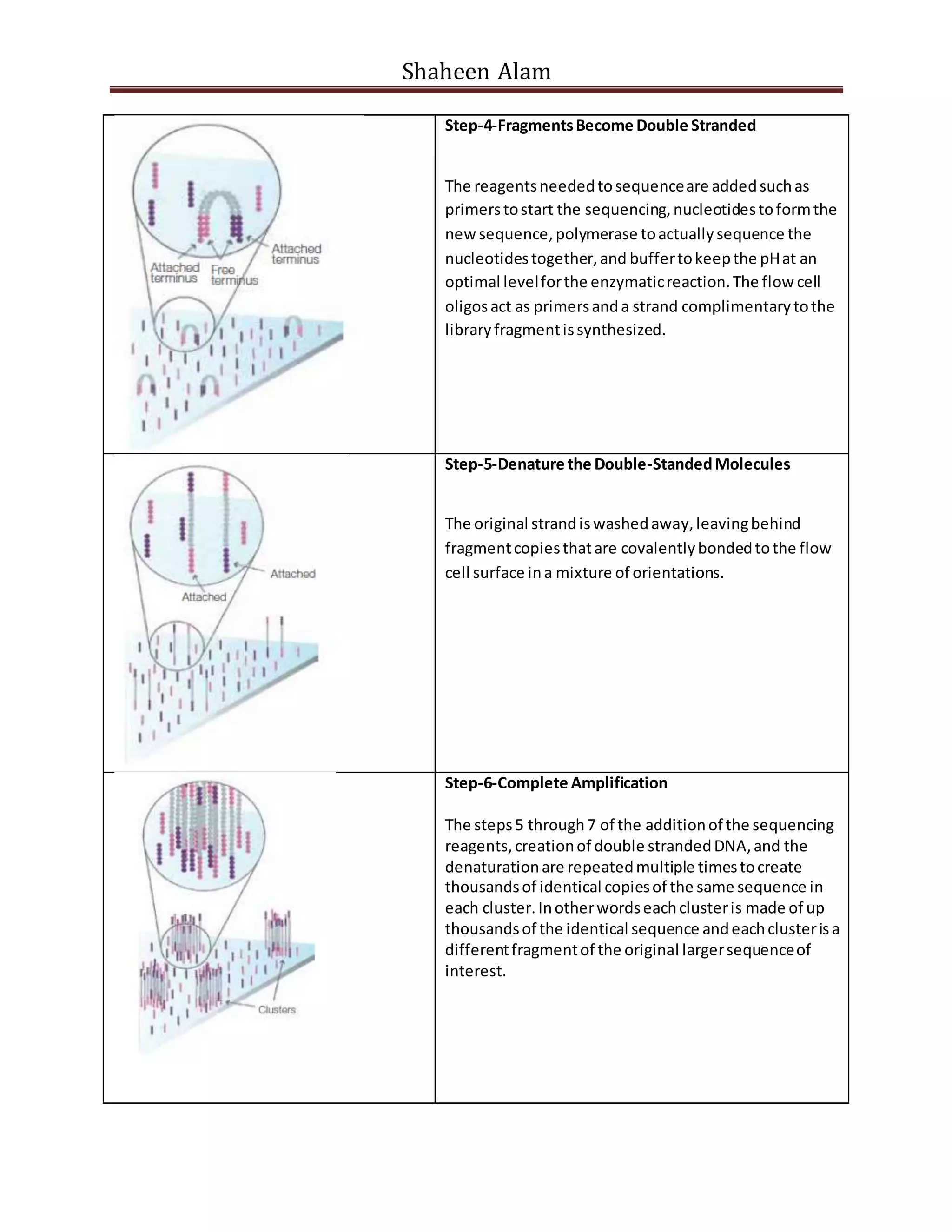
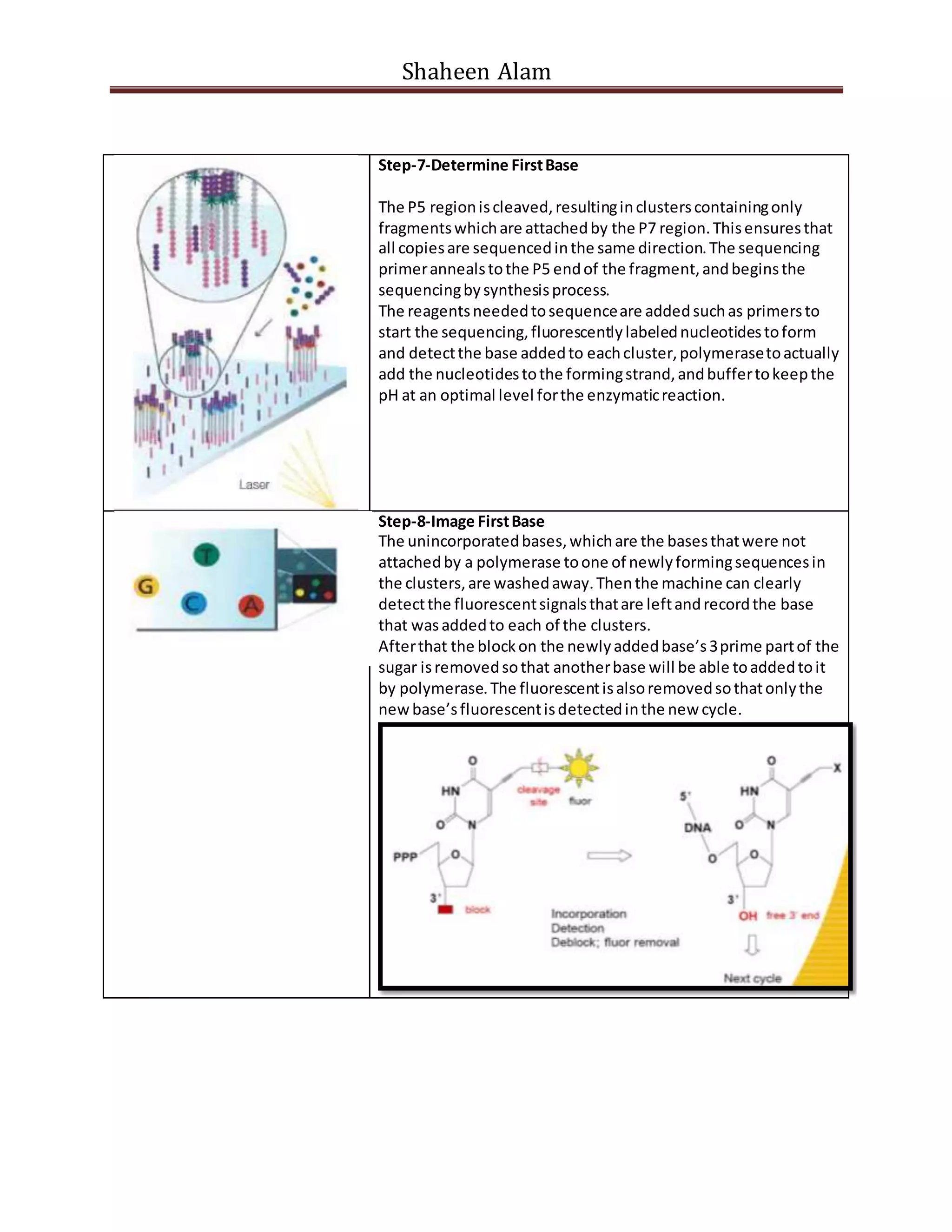
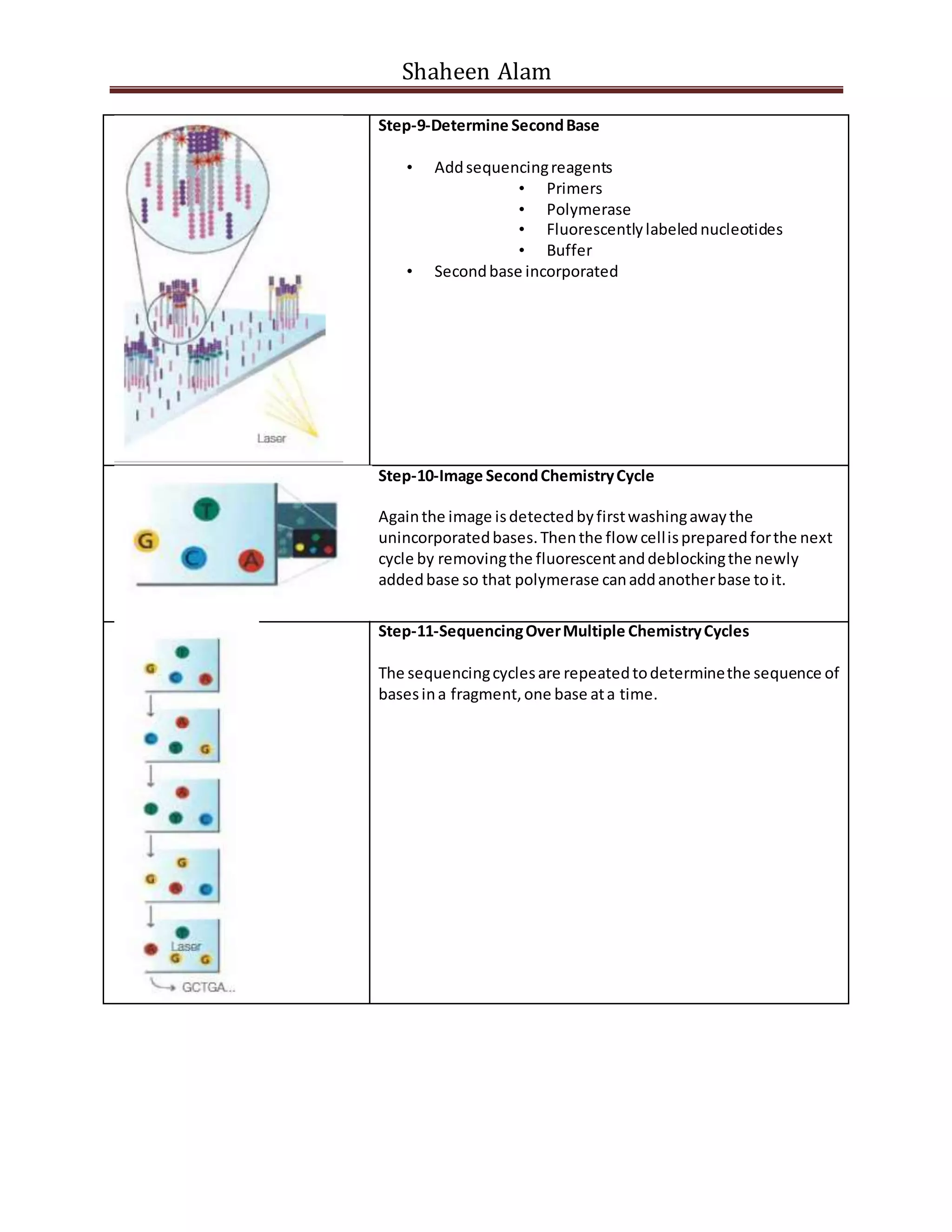
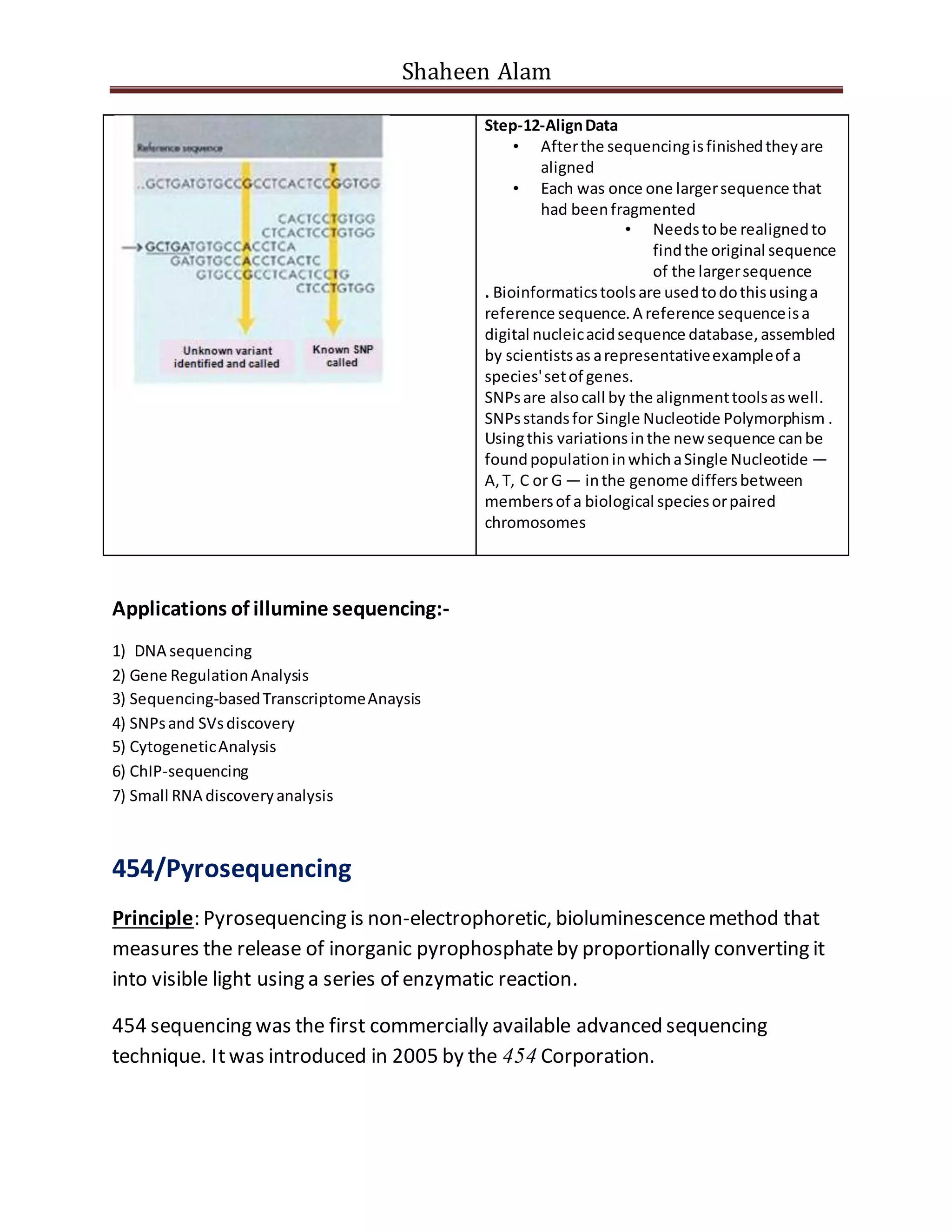
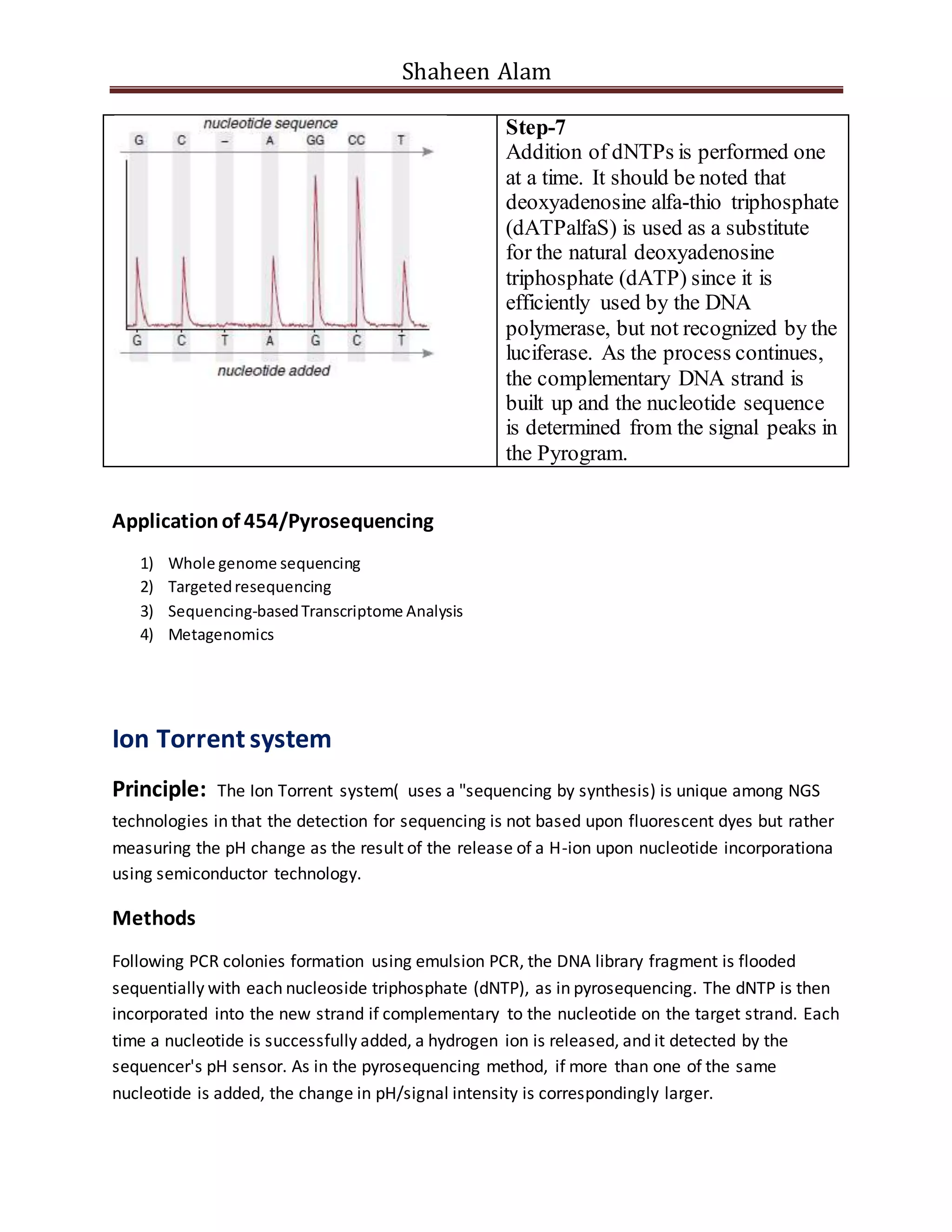
Next generation sequencing techniques allow for high-throughput DNA sequencing at a lower cost compared to Sanger sequencing. The document focuses on Illumina sequencing and 454 pyrosequencing. In Illumina sequencing, DNA fragments are attached to a flow cell and undergo bridge amplification and sequencing by synthesis using fluorescently labeled nucleotides. 454 pyrosequencing involves emulsion PCR to amplify DNA fragments attached to beads, followed by sequencing using DNA polymerase and a bioluminescent detection of incorporated nucleotides. Both techniques allow for massively parallel sequencing of millions of DNA fragments.