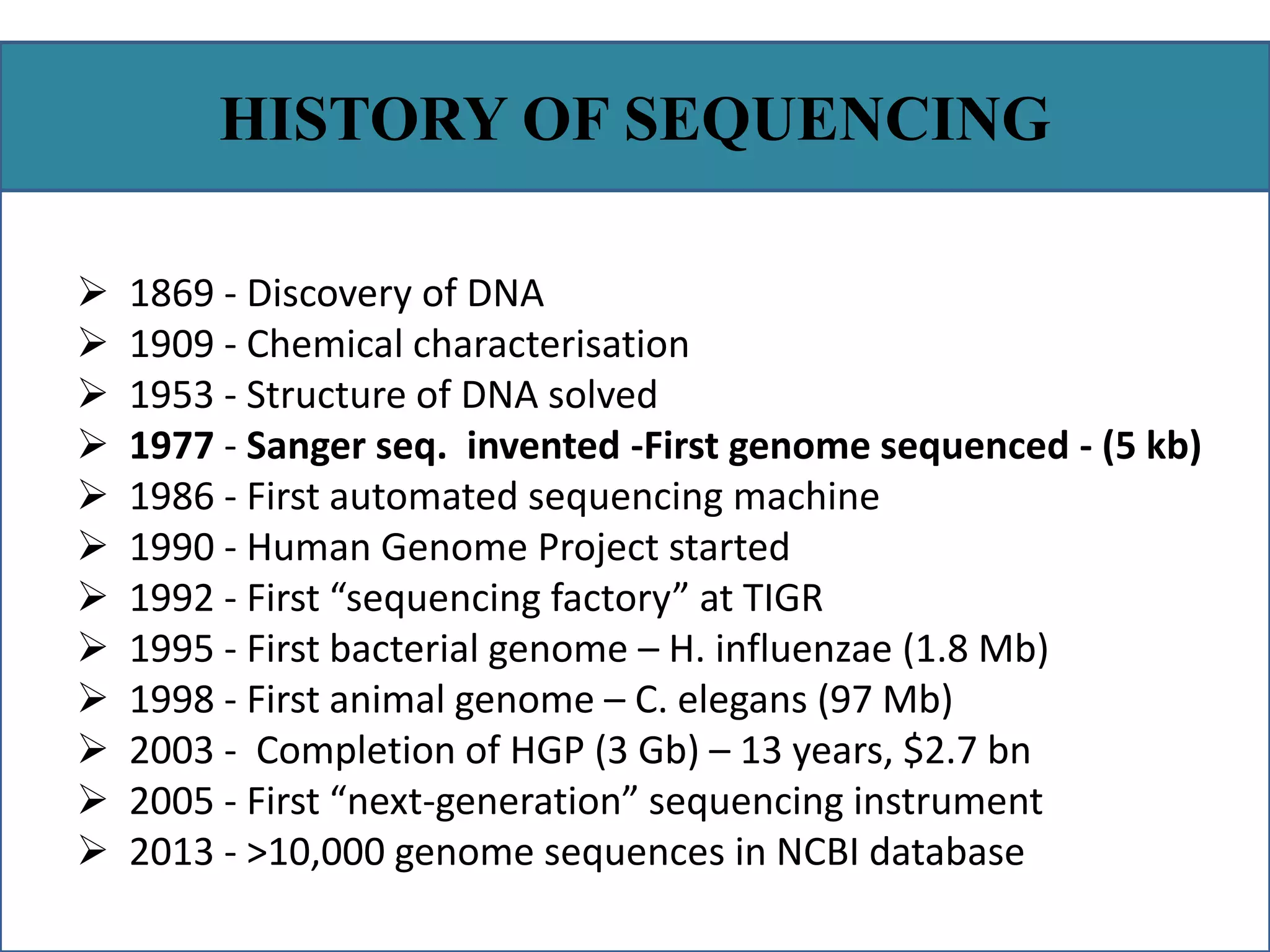
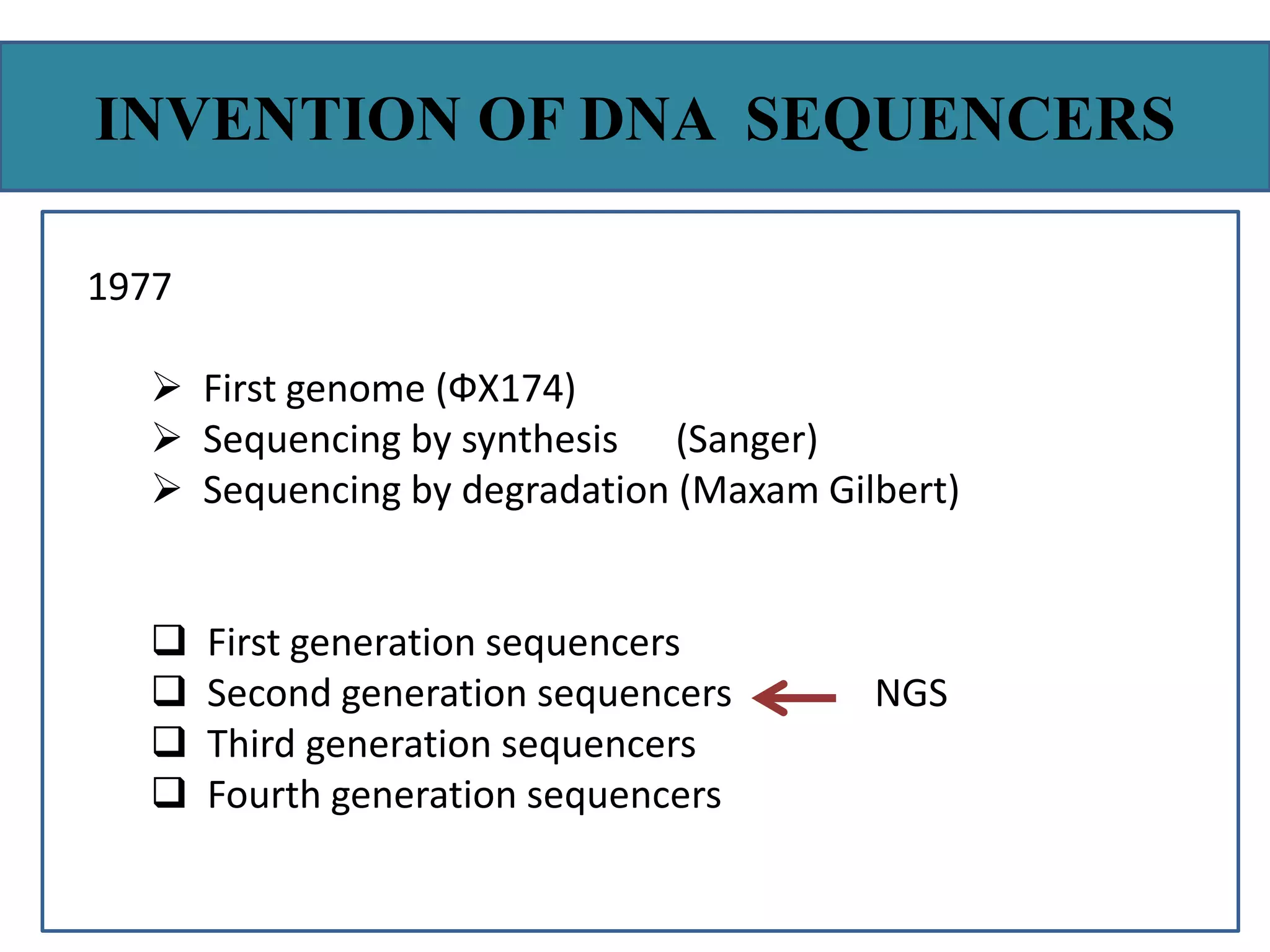
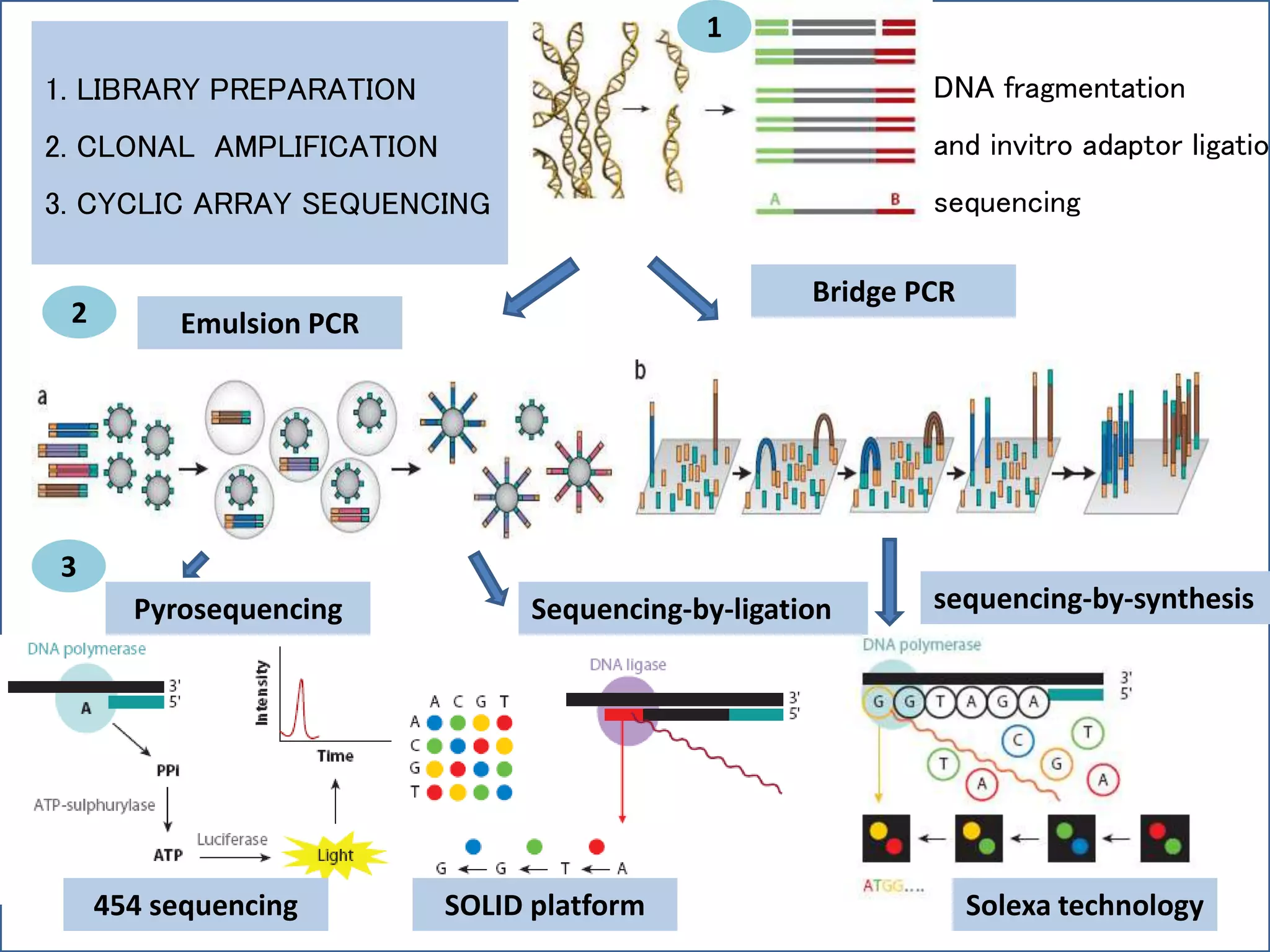
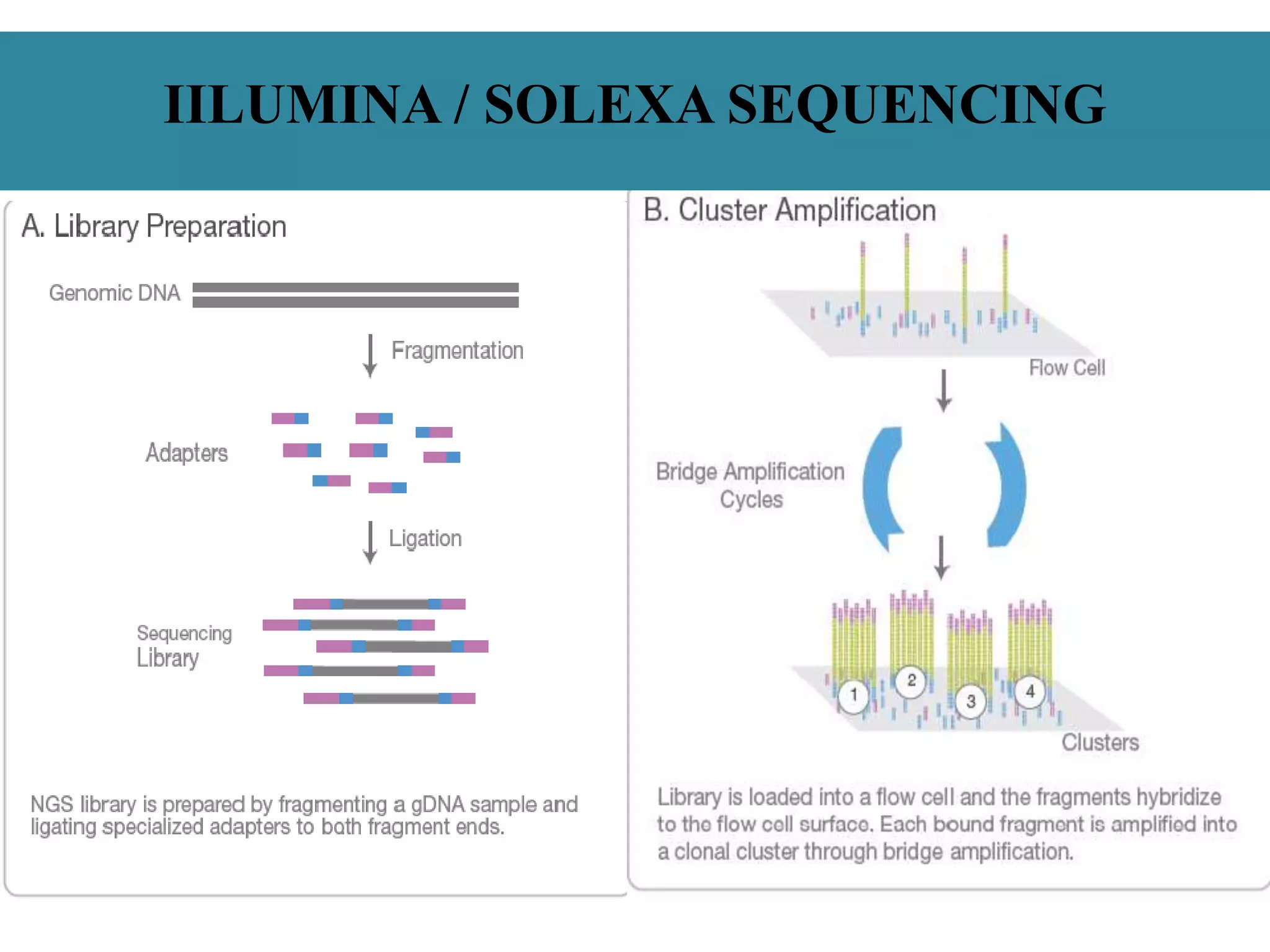
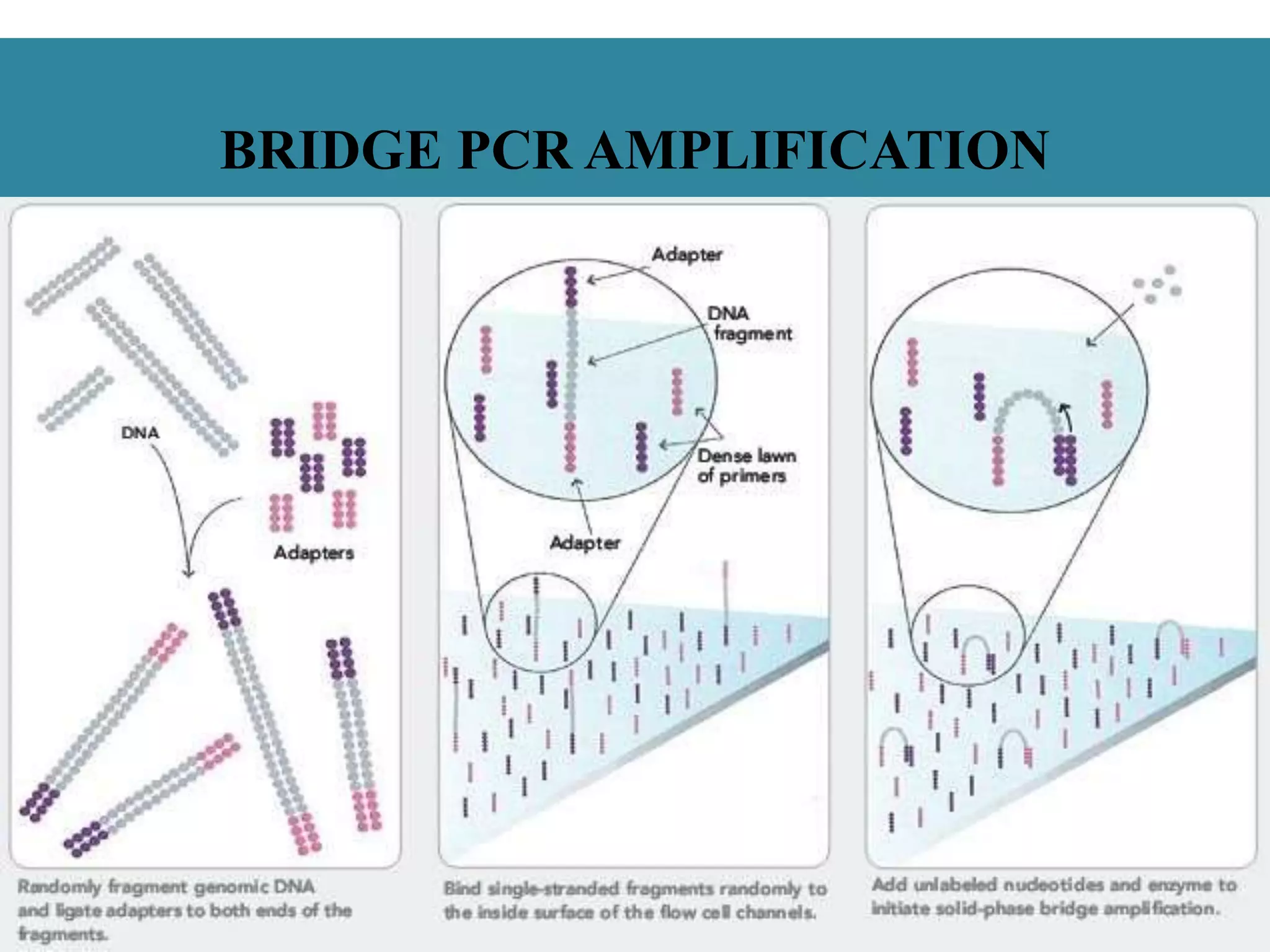
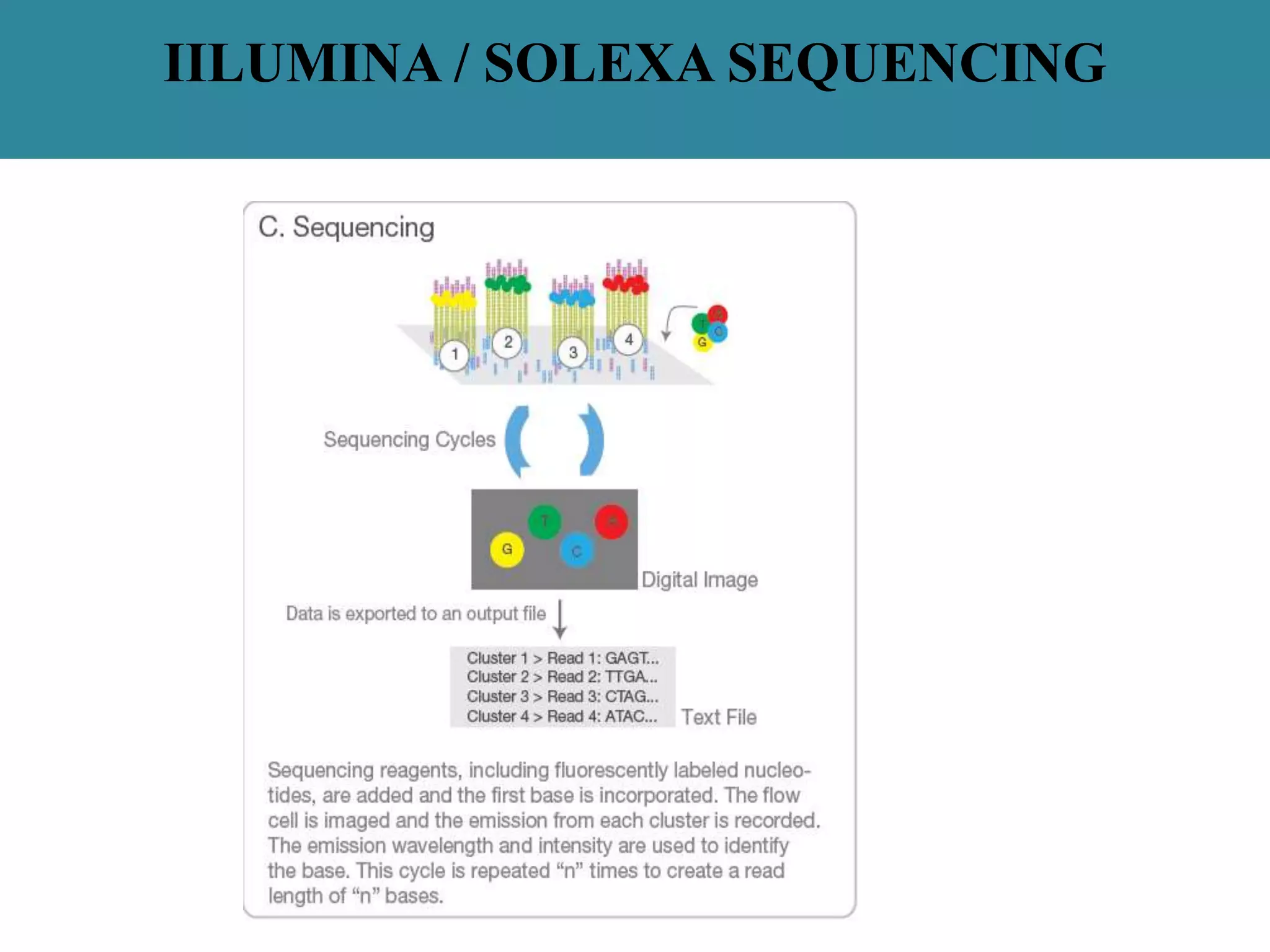
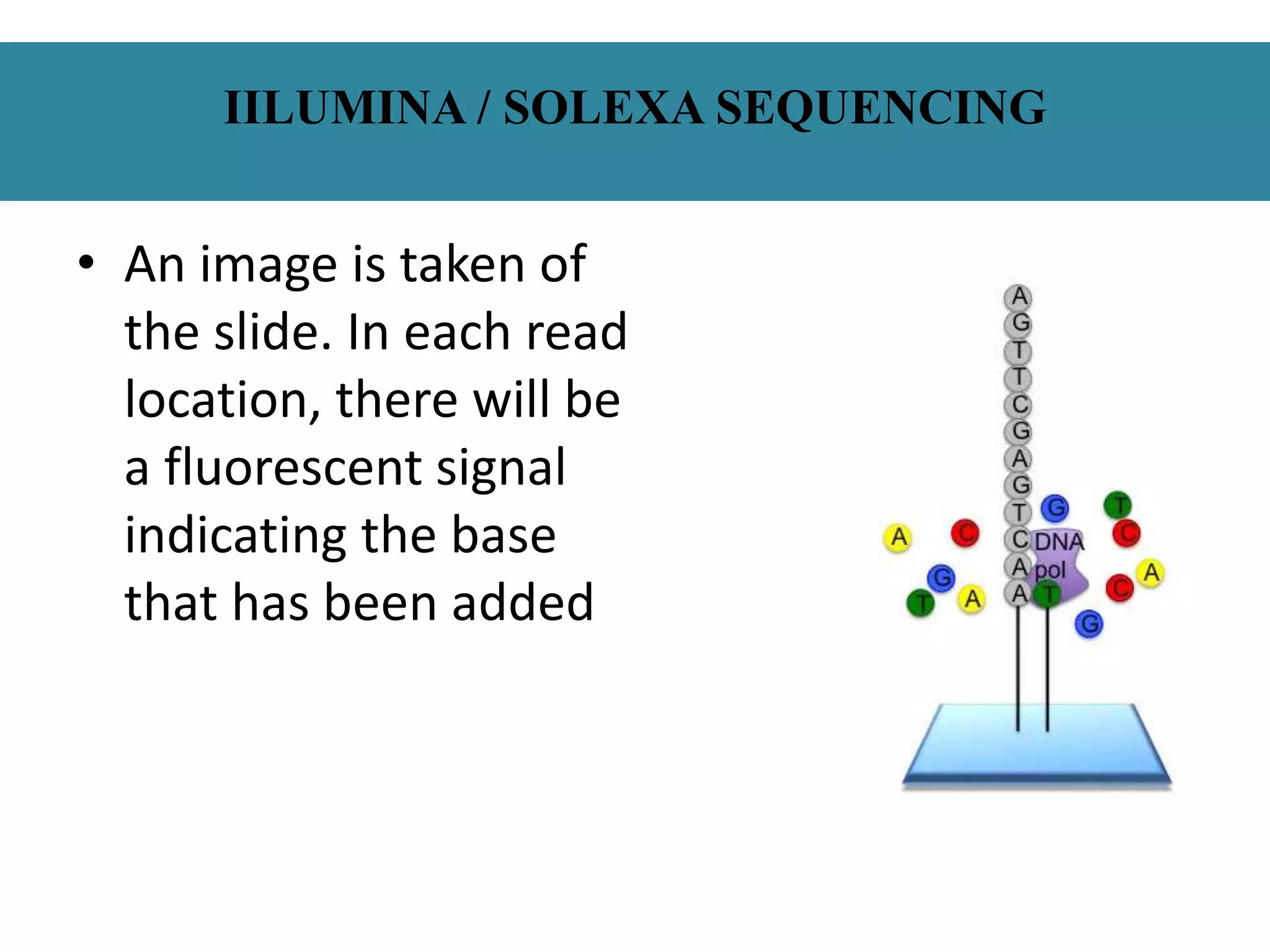
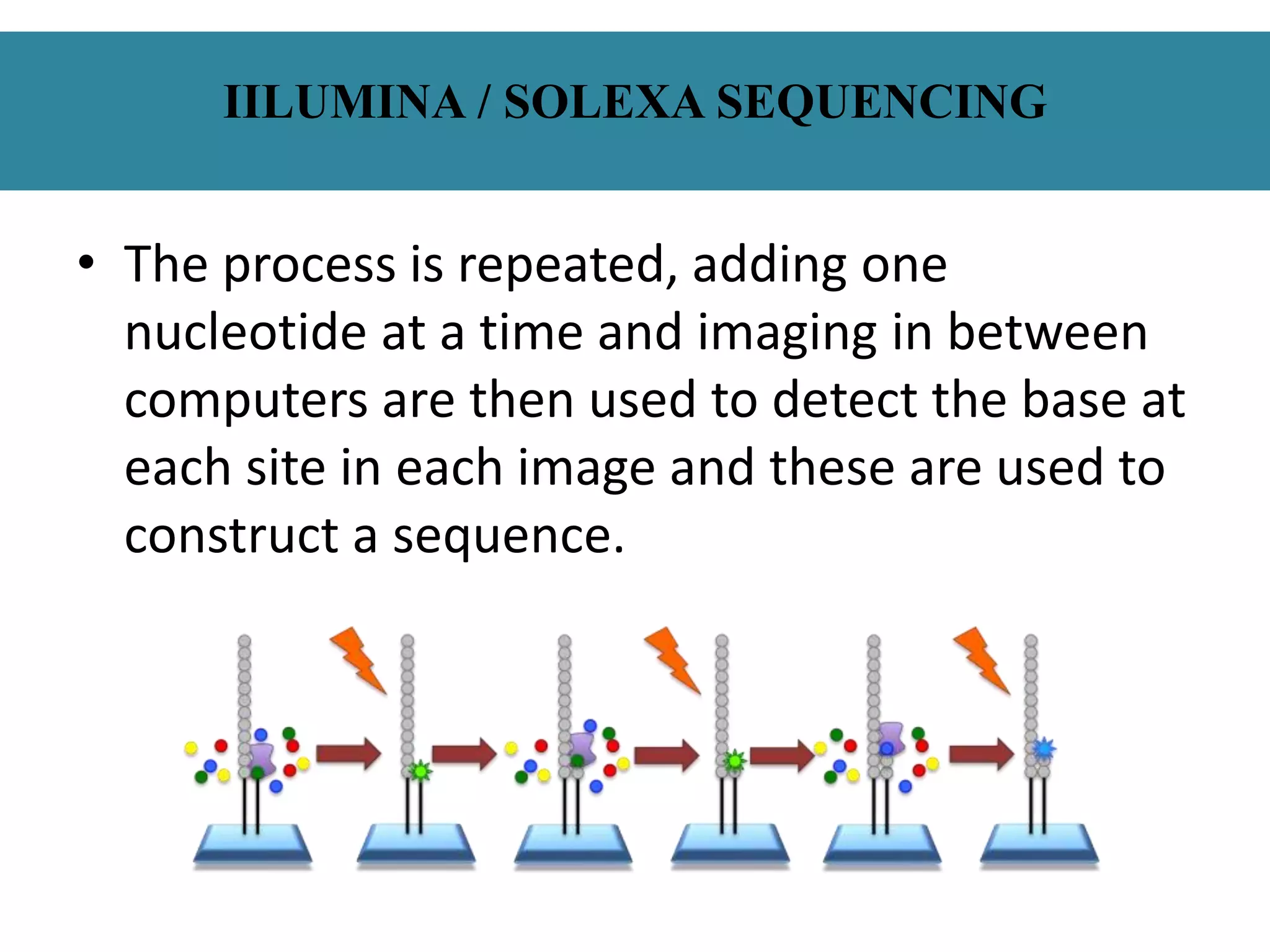
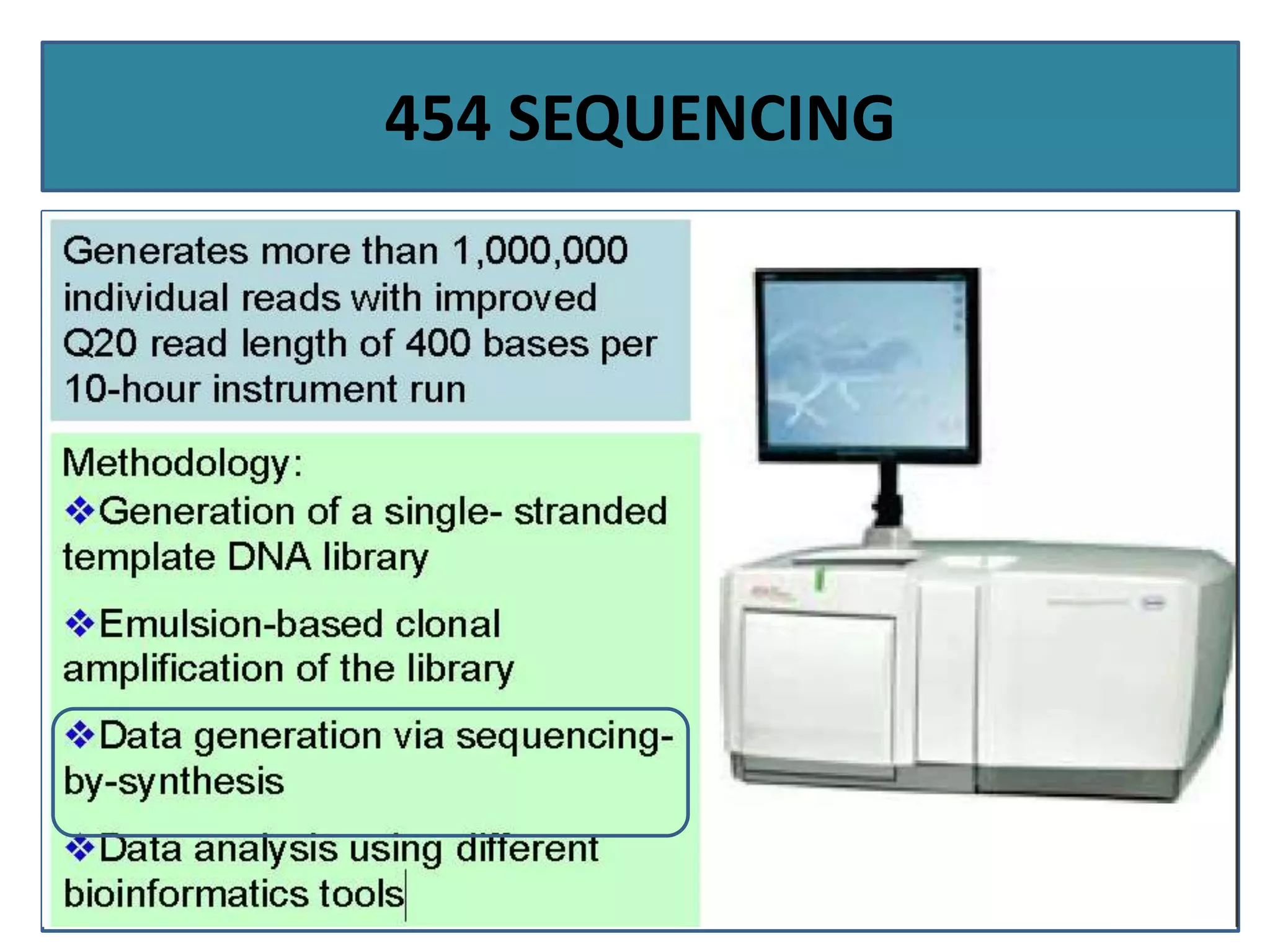
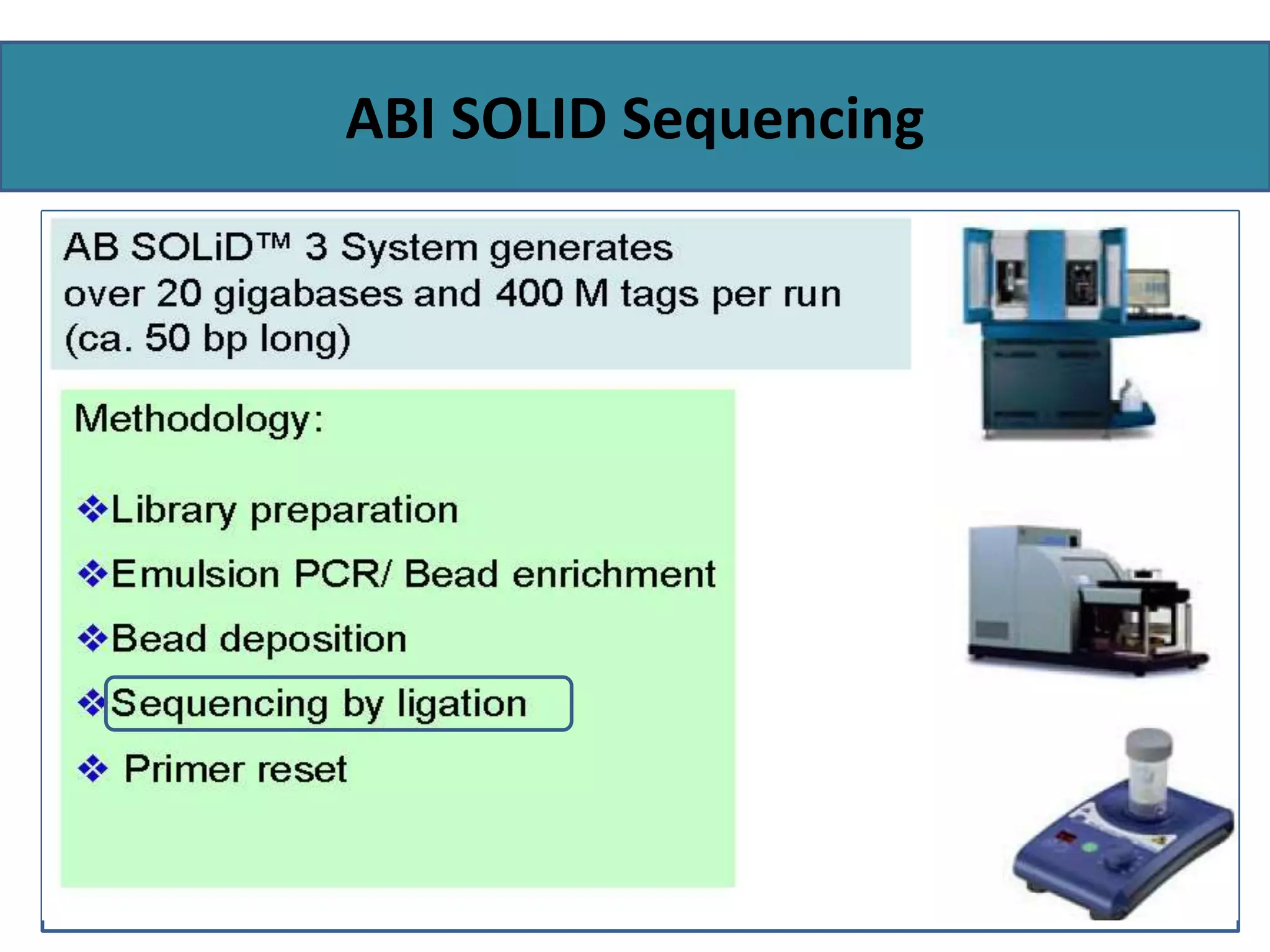
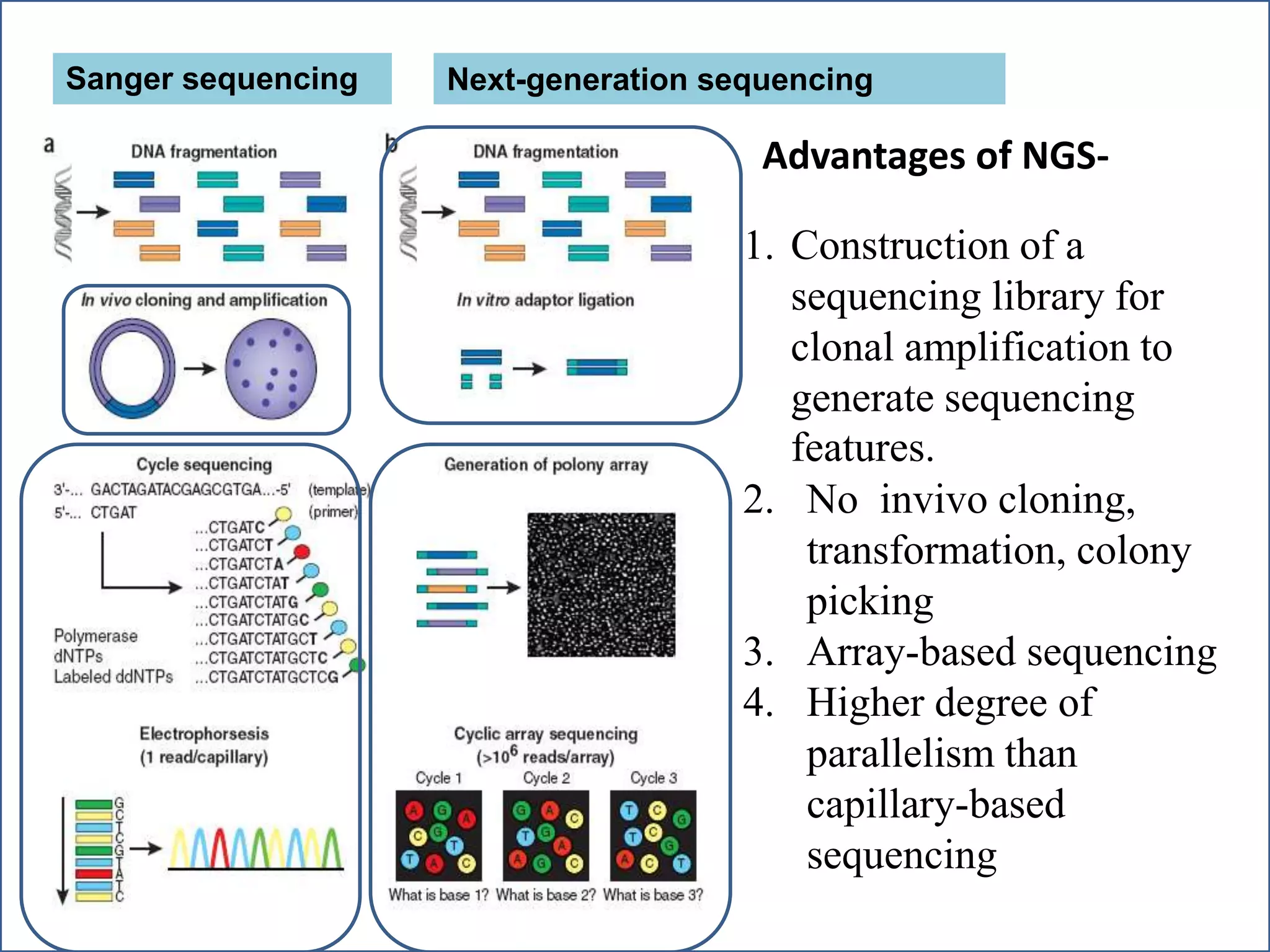
This document summarizes a class seminar presentation on next generation sequencing technologies. It begins with an overview of DNA sequencing and its importance. It then reviews the history of sequencing technologies, from the discovery of DNA to the development of Sanger sequencing and next generation sequencing platforms. The document focuses on describing the Illumina/Solexa, 454, and SOLiD next generation sequencing methods. It explains the key steps in library preparation, cluster amplification, and sequencing by synthesis or ligation for these platforms. The advantages of next generation sequencing technologies over Sanger sequencing are also highlighted.