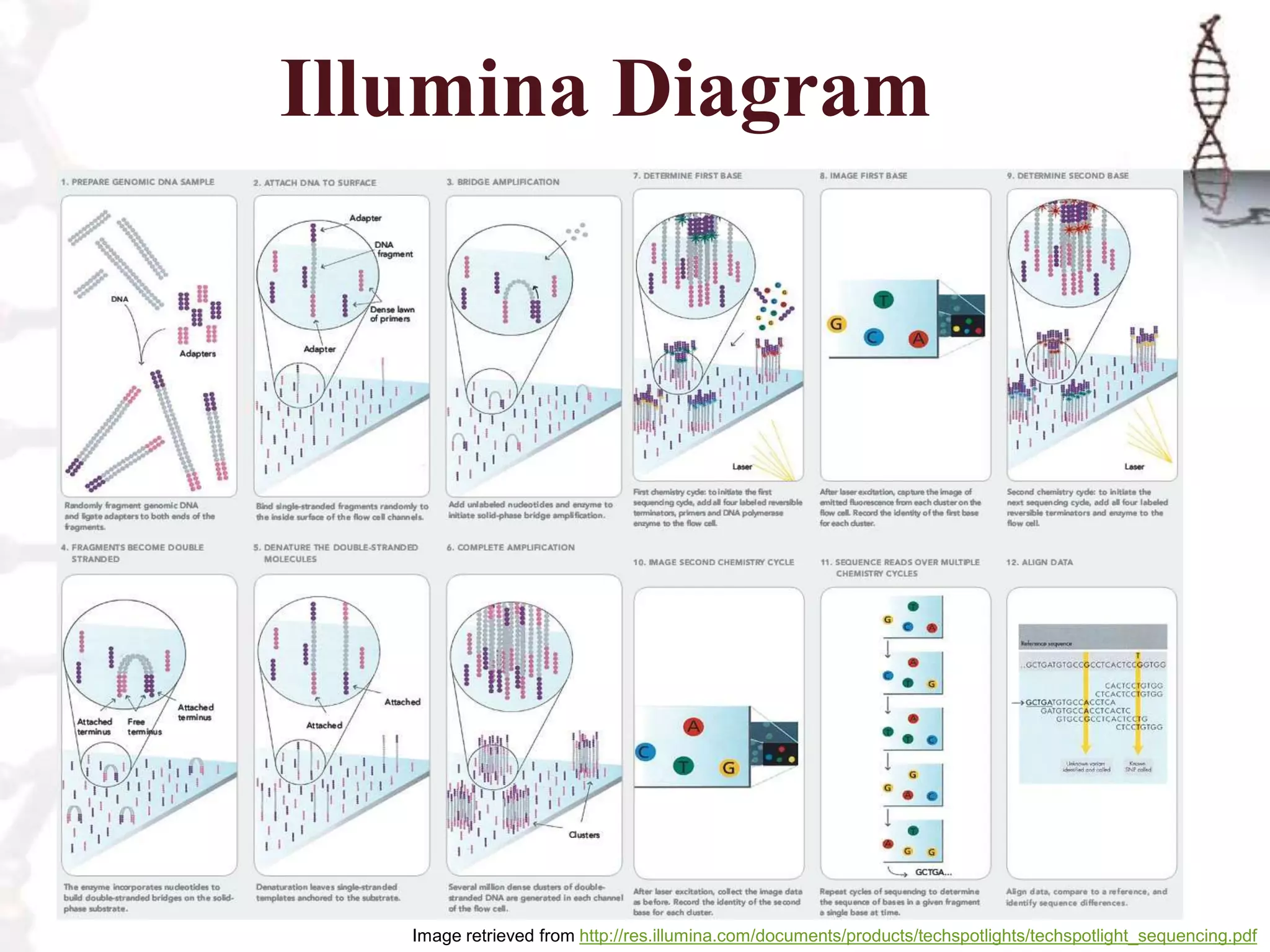
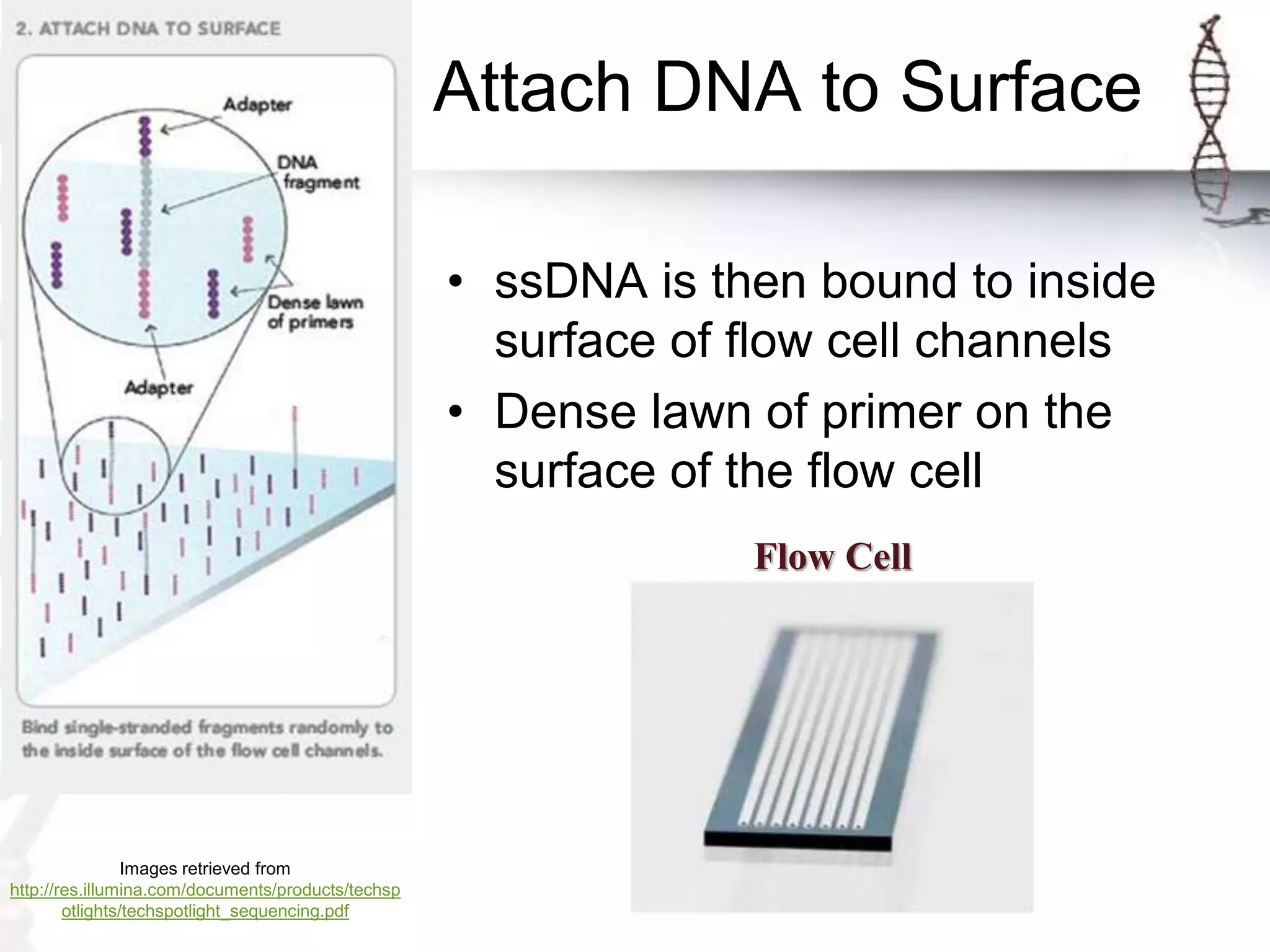
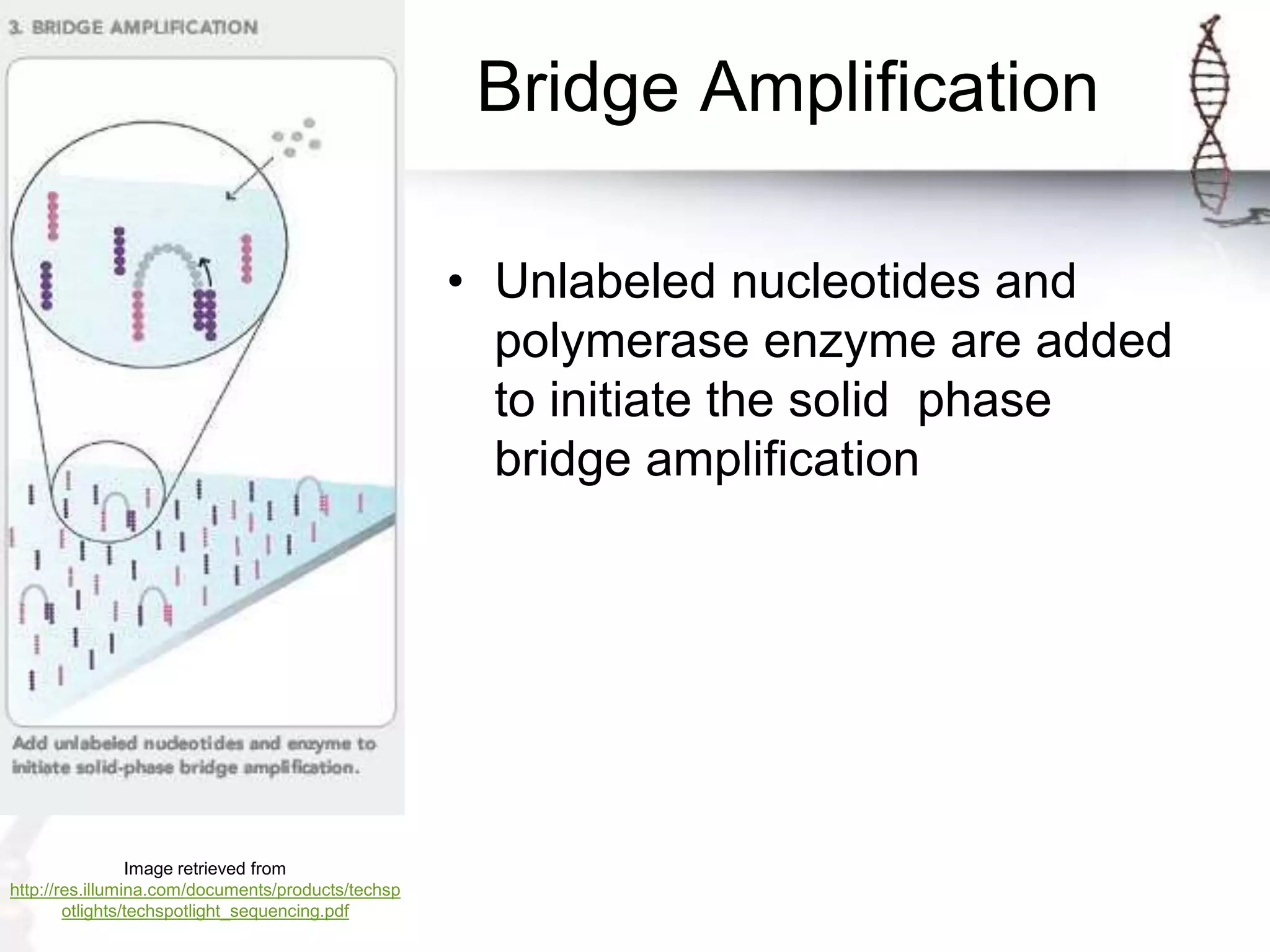
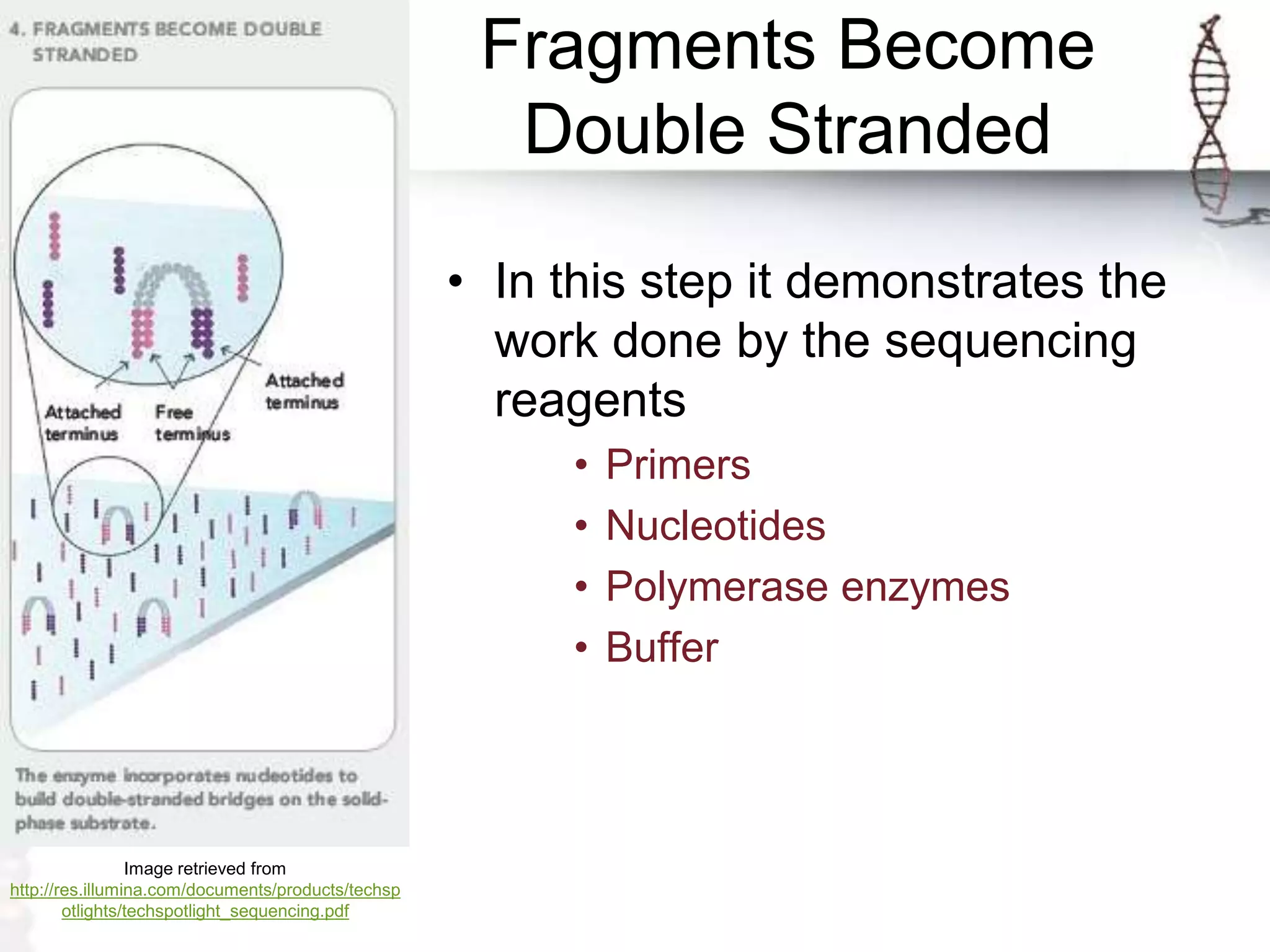
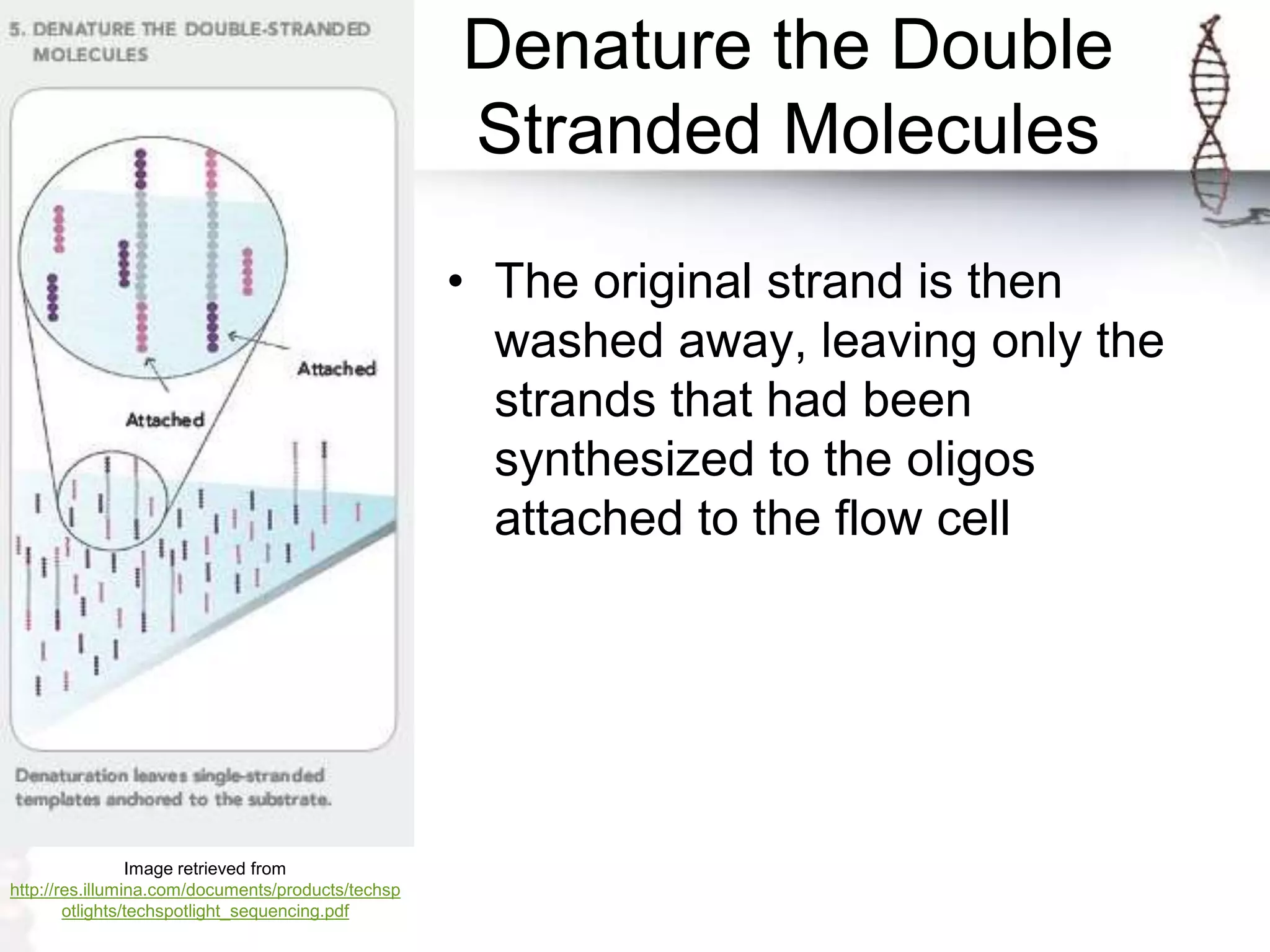
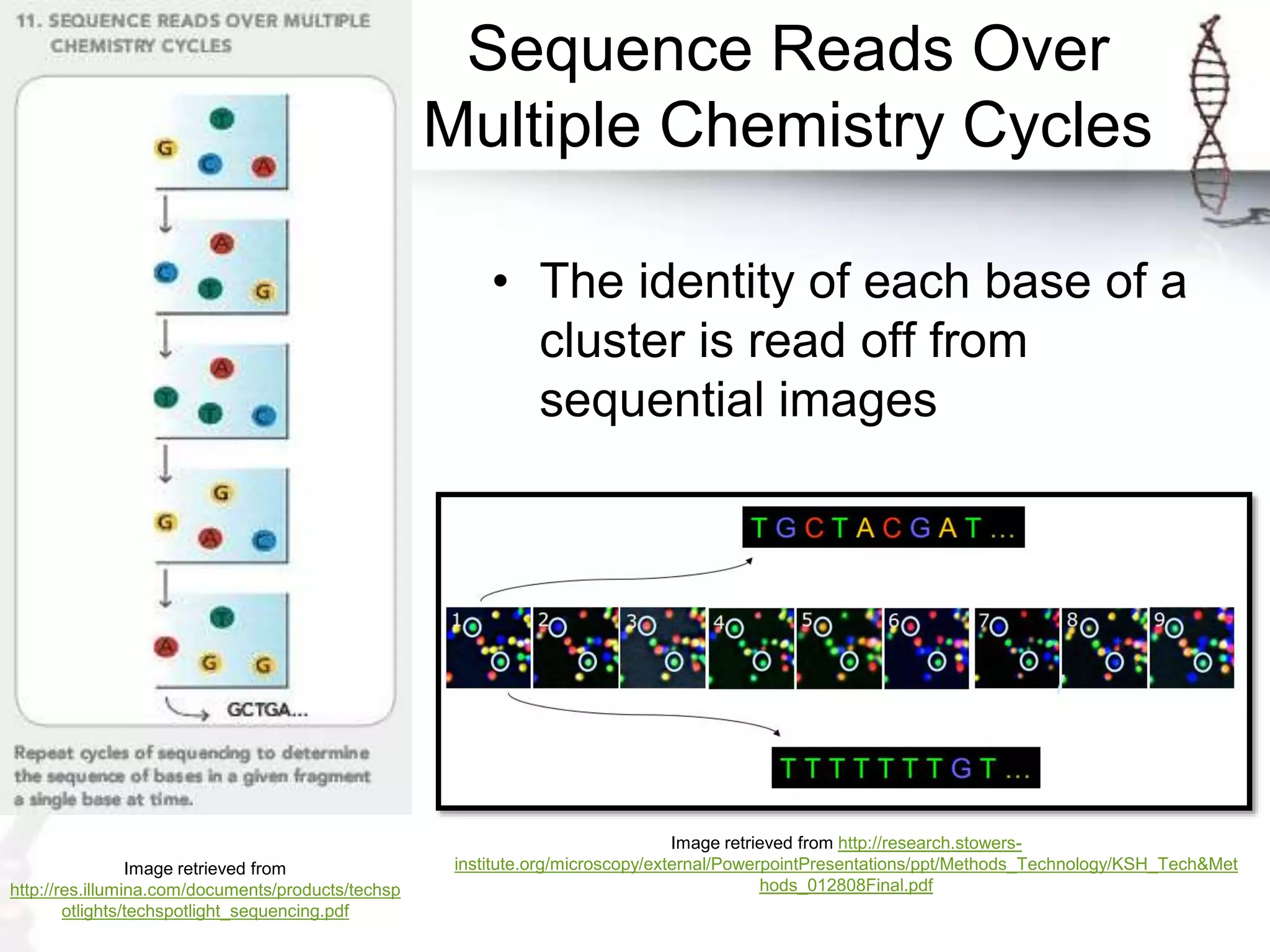
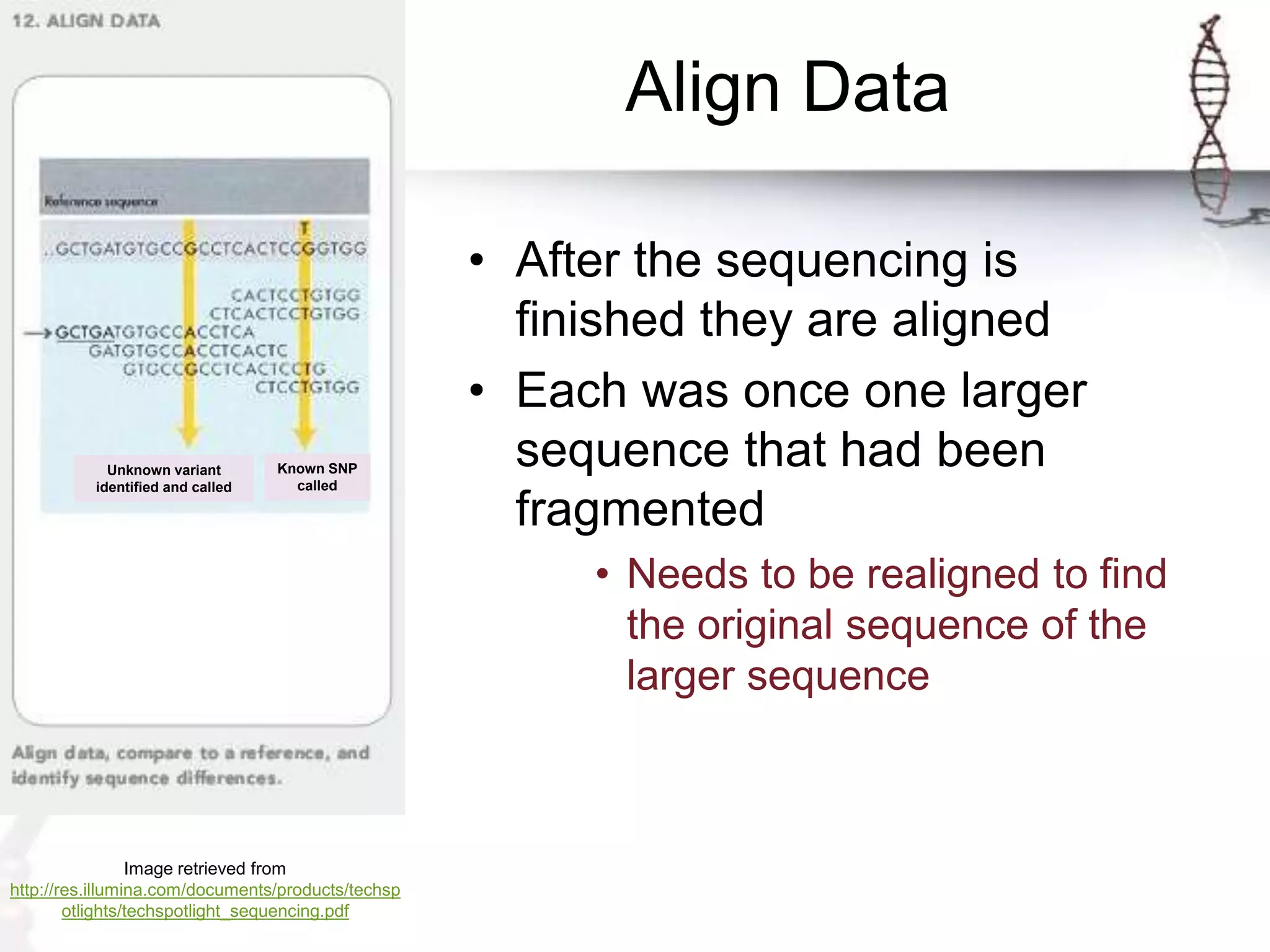
The document describes the steps of Illumina sequencing. Genomic DNA is first fragmented and adapters are ligated to create single-stranded DNA fragments. These fragments are attached to a flow cell and undergo bridge amplification to create clusters of identical DNA fragments. Sequencing occurs through cycles of reversible terminator-based sequencing using fluorescently labeled dNTPs, imaging of the fluorescence, and cleavage of the label and terminator to allow the next cycle. After multiple cycles, the sequenced reads are aligned to the reference genome to determine the original sequence.