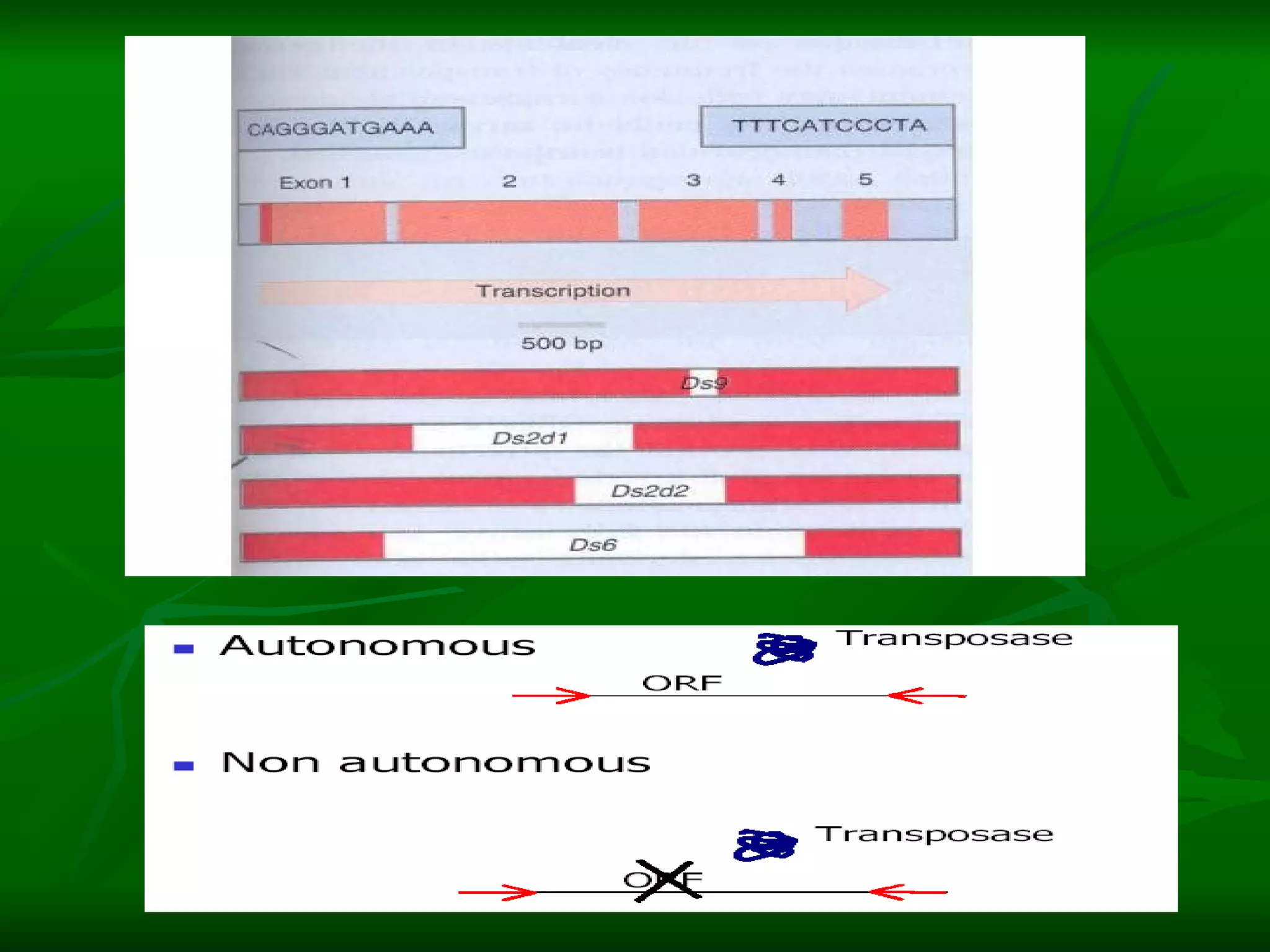
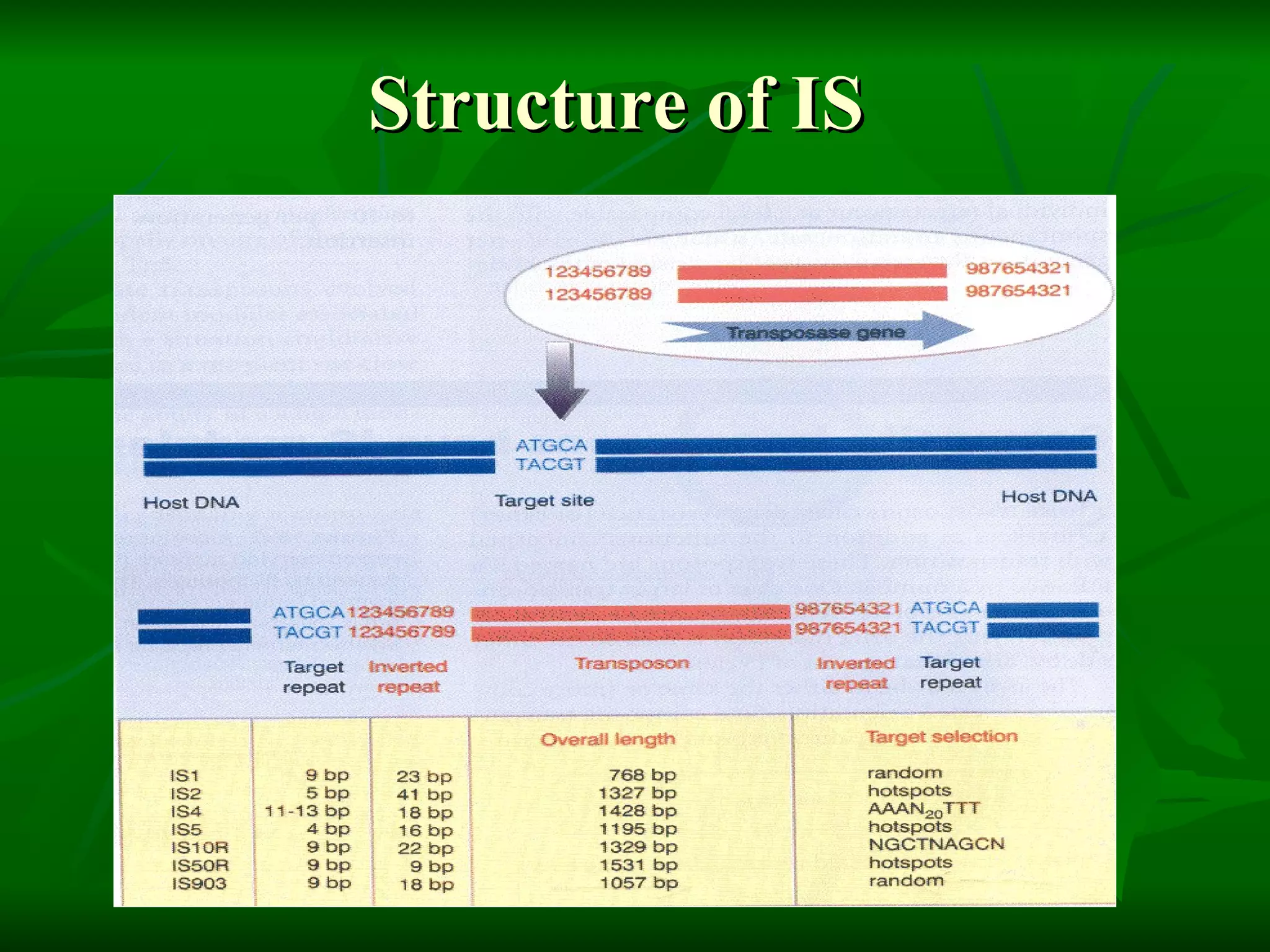
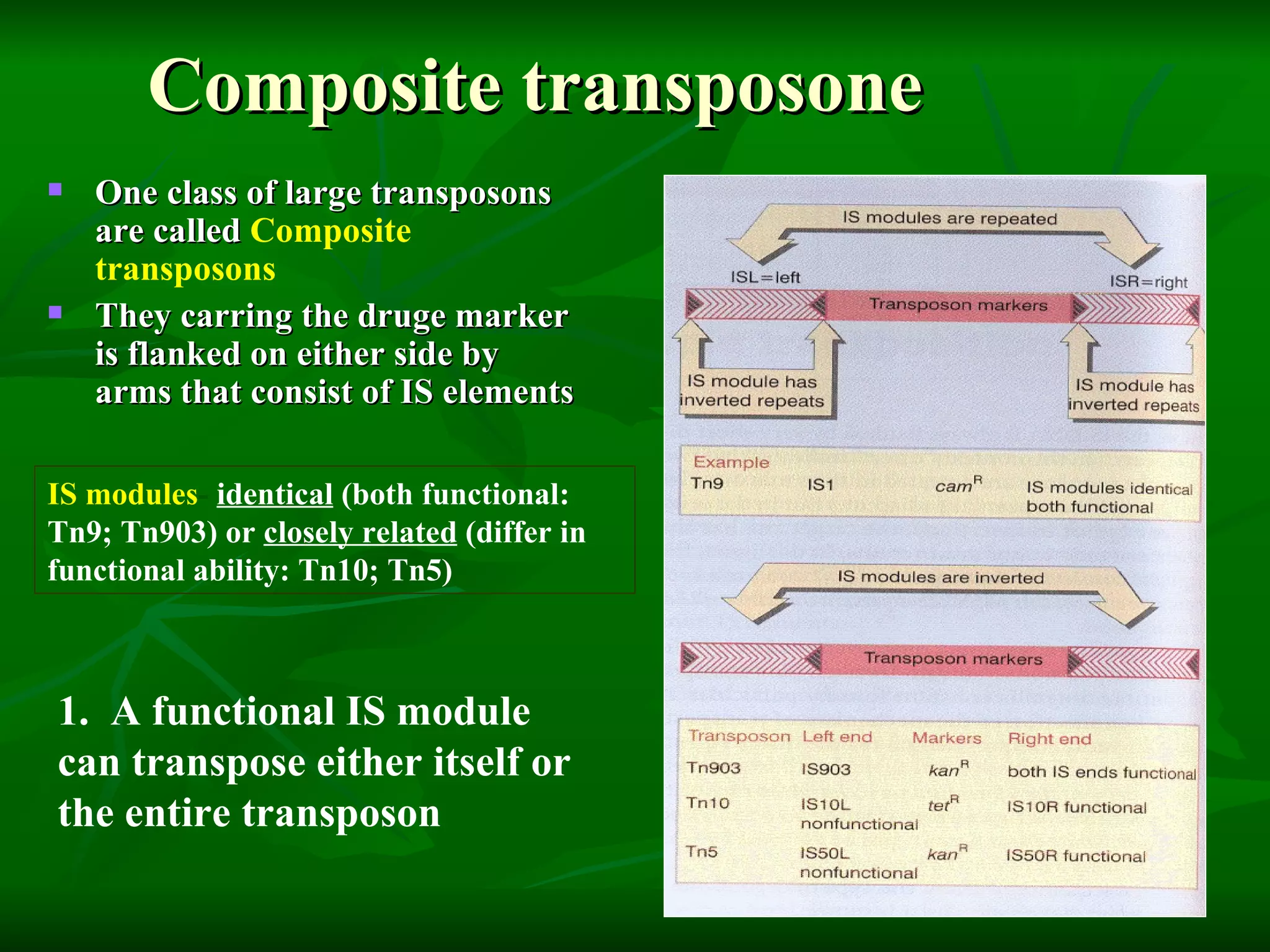
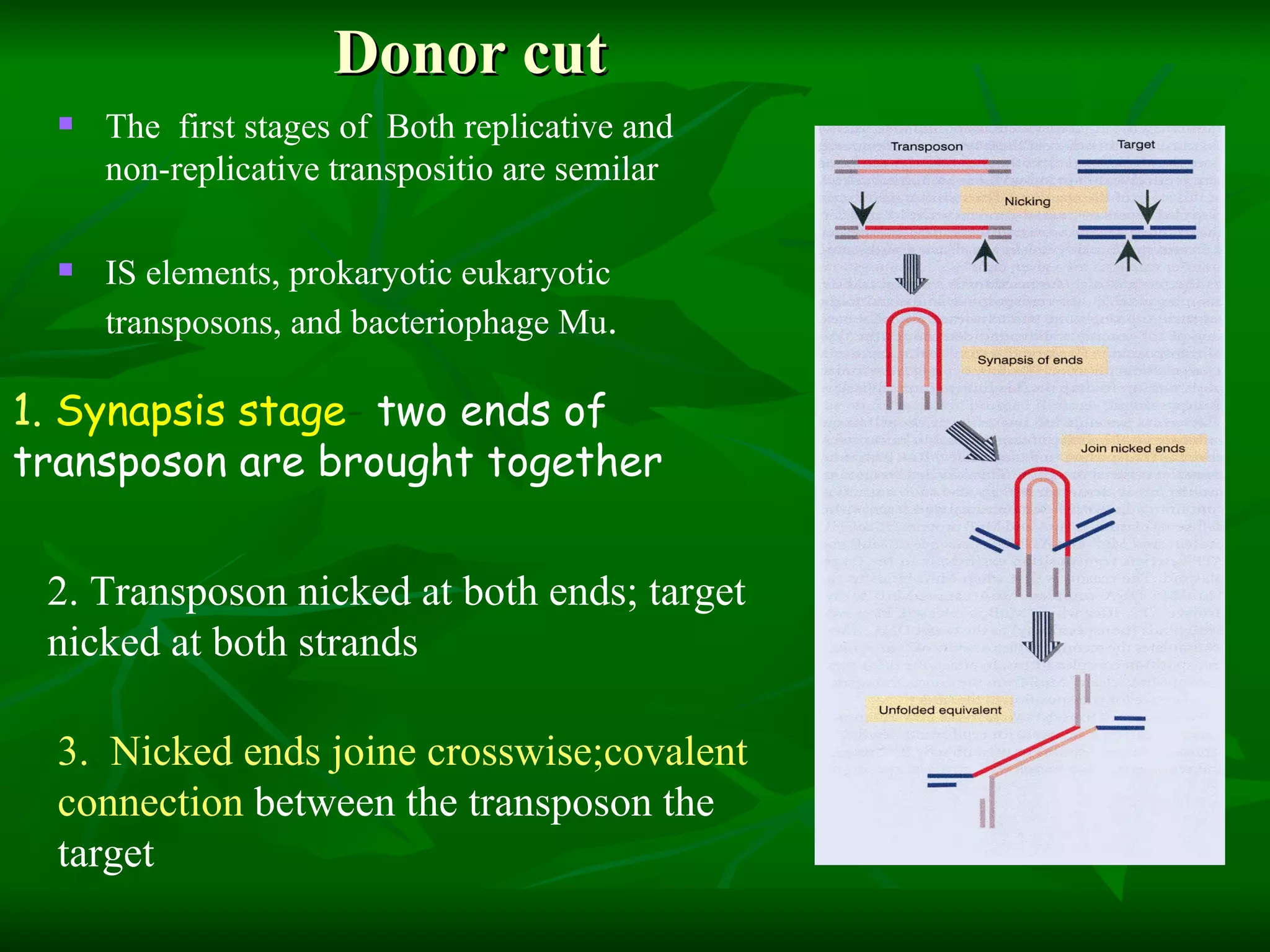
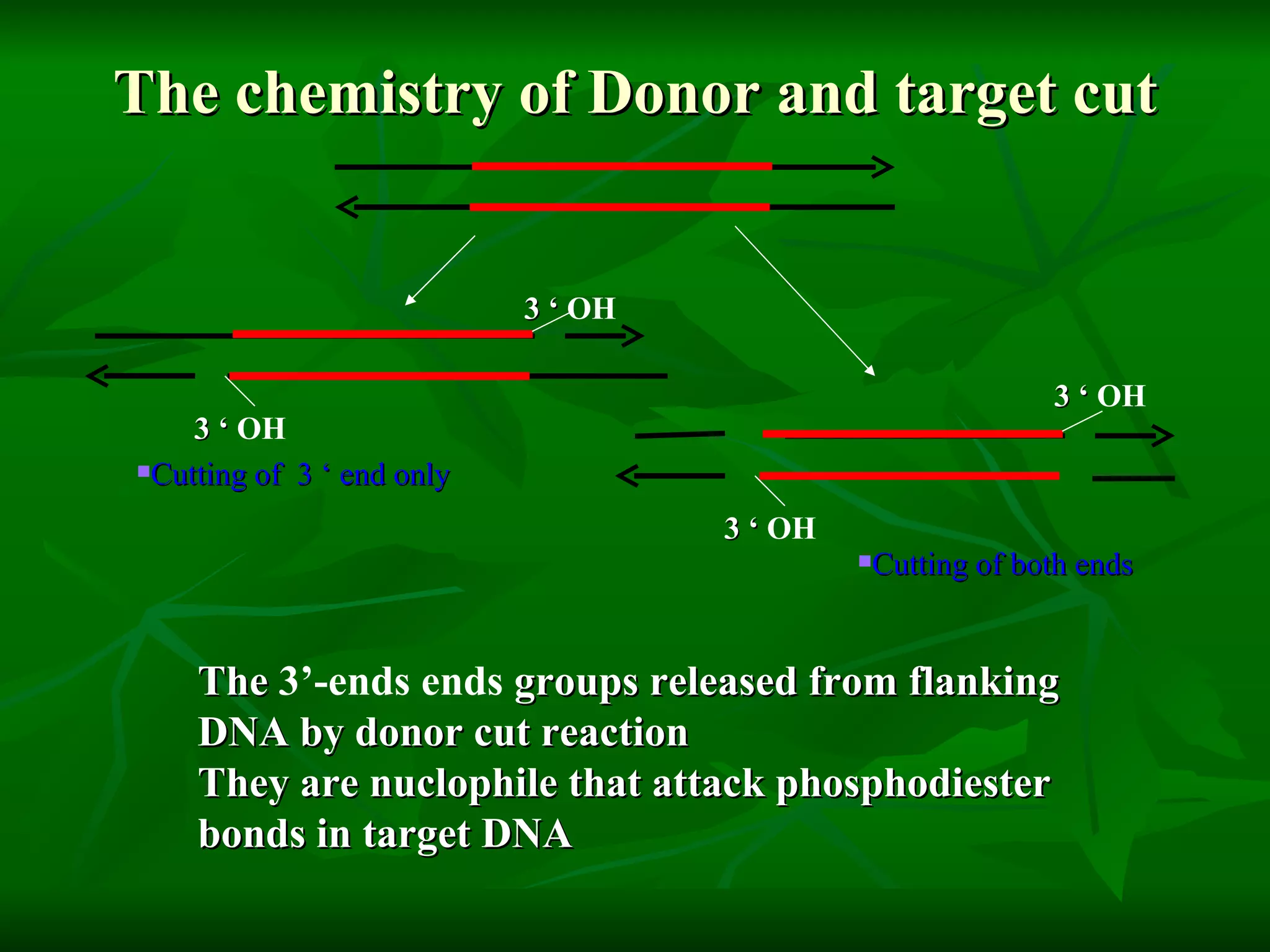
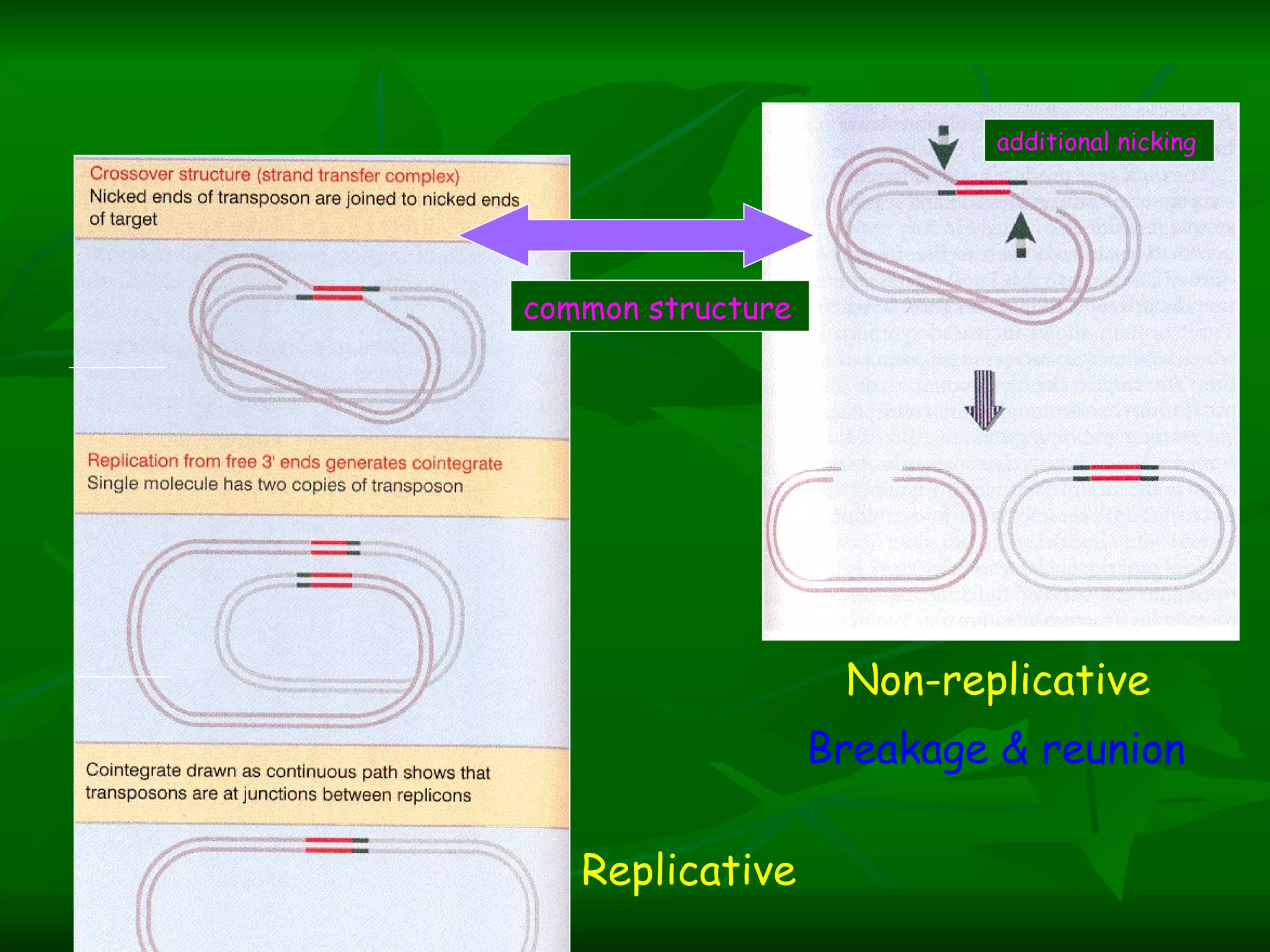
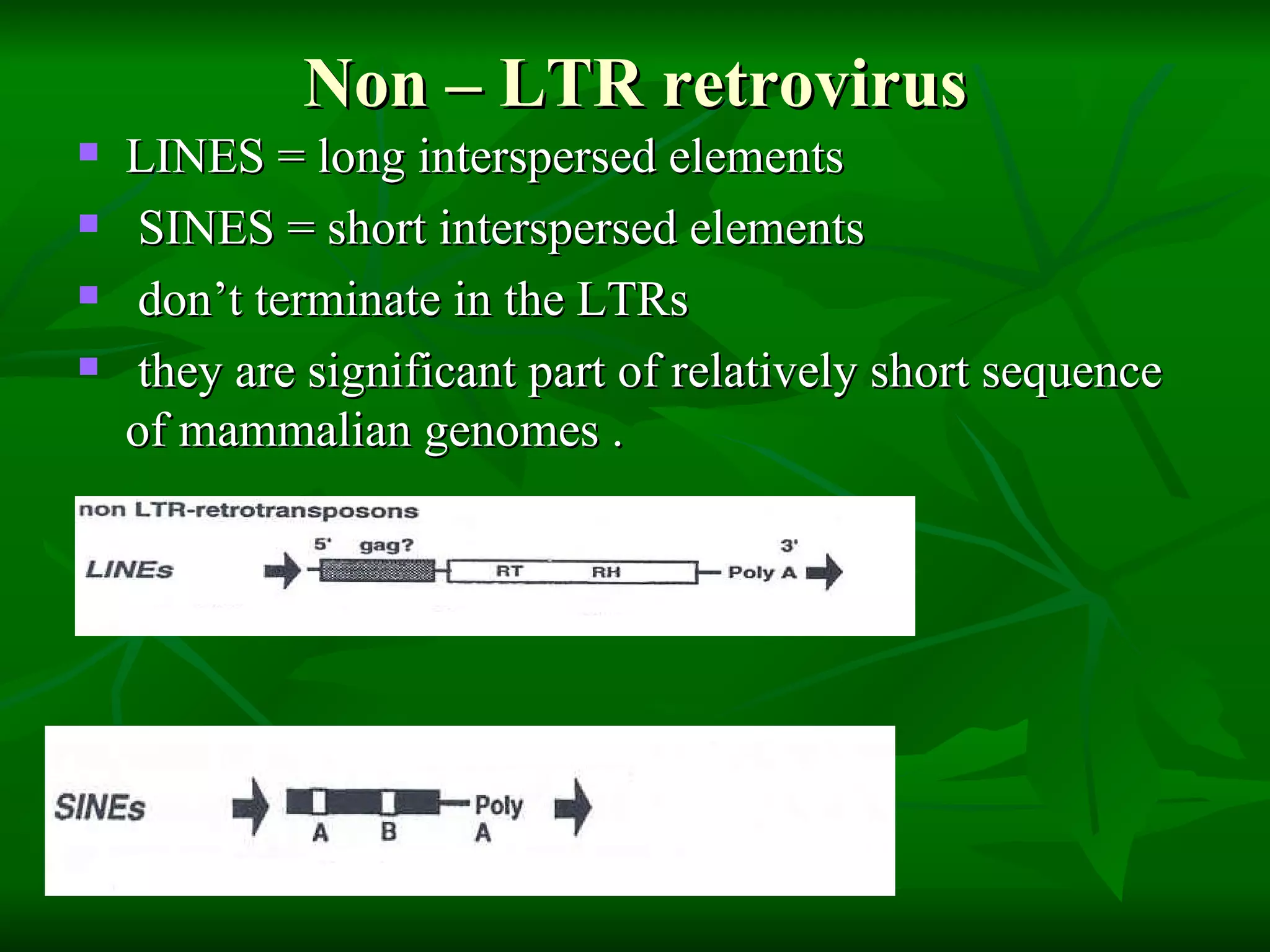
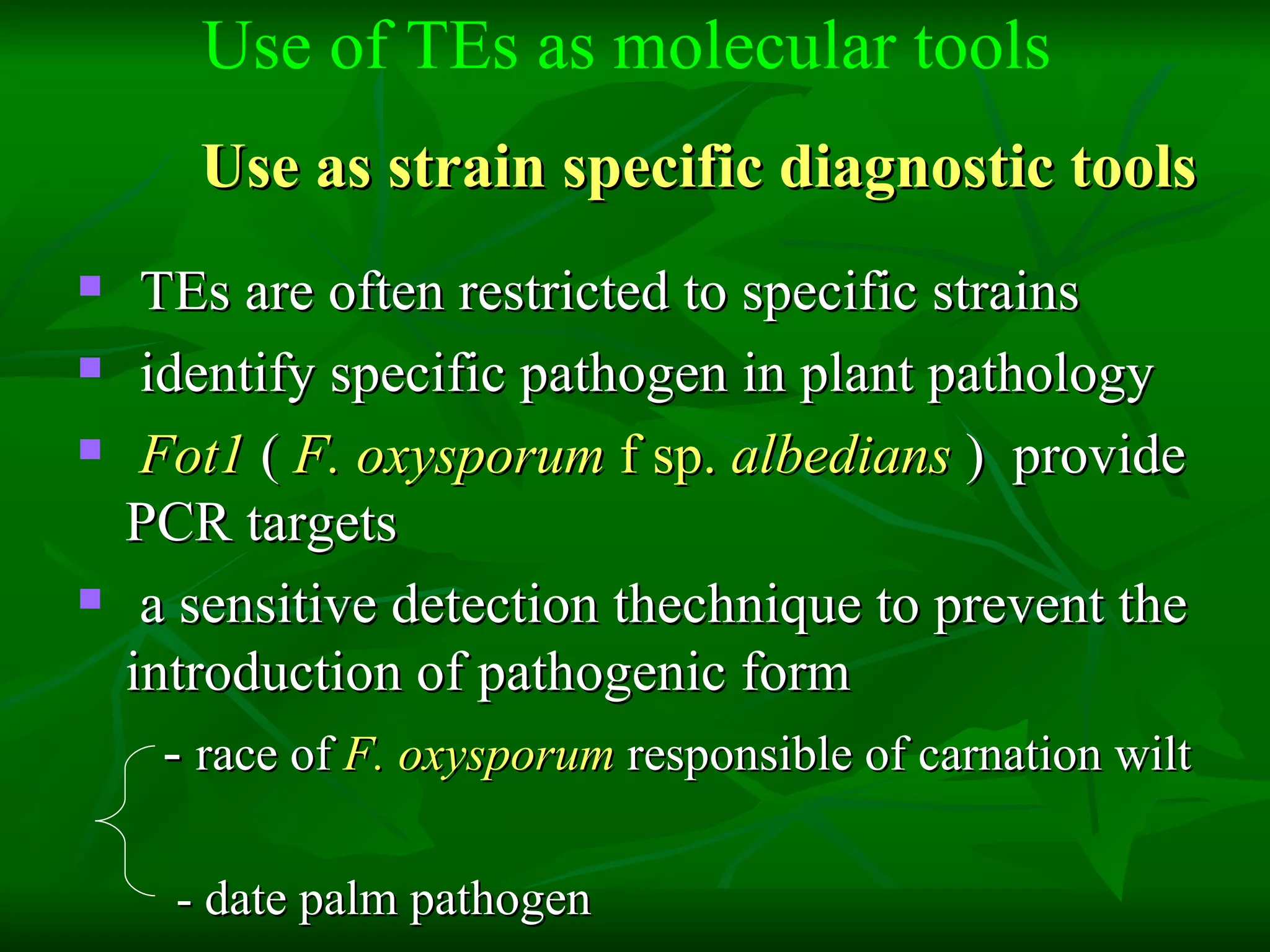
Transposable elements are segments of DNA that can move within genomes. They are present in all domains of life and have been shown to drive evolution by causing mutations through insertion, deletion, and rearrangement. Barbara McClintock discovered transposons in maize in the 1940s and was awarded a Nobel Prize for this work. Transposable elements are classified as DNA transposons or retrotransposons, and can be further divided into autonomous and non-autonomous types based on their ability to excise and transpose independently.