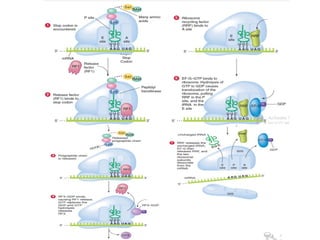
The document explains the process of translation, where mRNA is converted into amino acid sequences through the interactions of codons, anticodons, and ribosomes. Key characteristics of the genetic code include its triplet nature, lack of commas, universality, and degeneracy, with specific start and stop codons. The translation process involves initiation, elongation, and termination steps, facilitated by tRNA, ribosomal RNA, and various factors.